8PHI
 
 | | Crystal structure of prefusion-stabilized RSV F Variant DS-Cav1 in complex with Lonafarnib | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, CALCIUM ION, ... | | 著者 | Rajak, M, Krey, T. | | 登録日 | 2023-06-19 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.289 Å) | | 主引用文献 | Drug repurposing screen identifies lonafarnib as respiratory syncytial virus fusion protein inhibitor.
Nat Commun, 15, 2024
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YA0
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7EKQ
 
 | | CrClpP-S2c | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | 登録日 | 2021-04-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKO
 
 | | CrClpP-S1 | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | 著者 | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | 登録日 | 2021-04-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
6GH5
 
 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme transcription open complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Zhang, X.D. | | 登録日 | 2018-05-04 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
6W3J
 
 | | Crystal structure of the FAM46C/Plk4/Cep192 complex | | 分子名称: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4, Terminal nucleotidyltransferase 5C | | 著者 | Chen, H, Lu, D.F, Shang, G.J, Zhang, X.W. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (4.385 Å) | | 主引用文献 | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
5L2Z
 
 | | Factor VIIa in complex with the inhibitor 1-[(2R,15R)-2-[(1-amino-4-fluoroisoquinolin-6-yl)amino]-4,15,17-trimethyl-3,12-dioxo-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaen-7-yl]cyclohexane-1-carboxylic acid | | 分子名称: | 1-[(2R,15R)-2-[(1-amino-4-fluoroisoquinolin-6-yl)amino]-4,15,17-trimethyl-3,12-dioxo-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaen-7-yl]cyclohexane-1-carboxylic acid, CALCIUM ION, Coagulation factor VII (Heavy Chain), ... | | 著者 | Wei, A. | | 登録日 | 2016-08-02 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Synthesis and P1' SAR exploration of potent macrocyclic tissue factor-factor VIIa inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5L2Y
 
 | | Factor VIIa in complex with the inhibitor 1-[(2R,15R)-2-[(1-amino-4-fluoroisoquinolin-6-yl)amino]-4,15,20-trimethyl-3,12-dioxo-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6,8,10(21),16,19-hexaen-7-yl] cyclobutane-1-carboxylic acid | | 分子名称: | 1-[(2R,15R)-2-[(1-amino-4-fluoroisoquinolin-6-yl)amino]-4,15,17-trimethyl-3,12-dioxo-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaen-7-yl]cyclobutane-1-carboxylic acid, CALCIUM ION, Coagulation factor VII (Heavy Chain), ... | | 著者 | Wei, A. | | 登録日 | 2016-08-02 | | 公開日 | 2016-09-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Synthesis and P1' SAR exploration of potent macrocyclic tissue factor-factor VIIa inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5SZX
 
 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | 著者 | Hong, S, Horton, J.R, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
5T0U
 
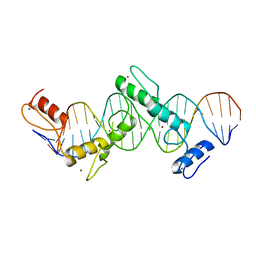 | | CTCF ZnF2-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-16 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.199 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5T01
 
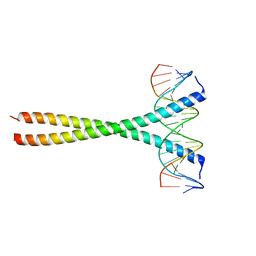 | | Human c-Jun DNA binding domain homodimer in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*AP*TP*GP*GP*AP*(5CM)P*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*GP*TP*CP*CP*AP*T)-3'), Transcription factor AP-1 | | 著者 | Hong, S, Horton, J.R, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
5LZ9
 
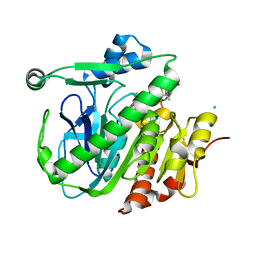 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | 分子名称: | 2-fluoranyl-5-[2-[(4~{S})-4-[4-methyl-1,1-bis(oxidanylidene)thian-4-yl]-2-oxidanylidene-pyrrolidin-1-yl]ethoxy]benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase, ... | | 著者 | Woolford, A, Day, P. | | 登録日 | 2016-09-29 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
5LZ5
 
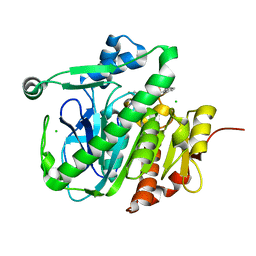 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | 分子名称: | 2-fluoranyl-5-[2-[(4~{S})-4-methyl-2-oxidanylidene-4-phenyl-pyrrolidin-1-yl]ethoxy]benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase | | 著者 | Woolford, A.J.A, Day, P.J. | | 登録日 | 2016-09-29 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
7W8S
 
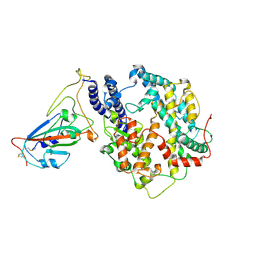 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Su, C, Qi, J.X, Gao, G.F. | | 登録日 | 2021-12-08 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
7WA1
 
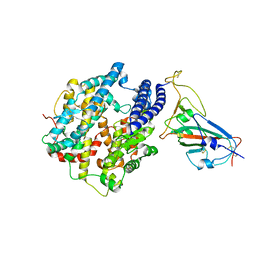 | |
5LZ7
 
 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | 分子名称: | 2-fluoranyl-5-[2-[(4~{R})-4-methyl-2-oxidanylidene-4-phenyl-pyrrolidin-1-yl]ethoxy]benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase | | 著者 | Woolford, A, Day, P. | | 登録日 | 2016-09-29 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | 著者 | Zhao, Z, Qi, J, Gao, F.G. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
5MSY
 
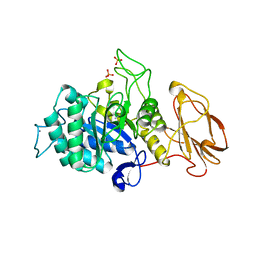 | | Glycoside hydrolase BT_1012 | | 分子名称: | AMMONIA, Glycoside hydrolase, PHOSPHATE ION | | 著者 | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | 登録日 | 2017-01-06 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQO
 
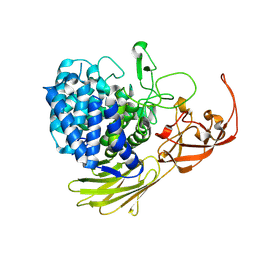 | | Glycoside hydrolase BT_1003 | | 分子名称: | Non-reducing end beta-L-arabinofuranosidase | | 著者 | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQS
 
 | | Sialidase BT_1020 | | 分子名称: | Beta-L-arabinobiosidase, CALCIUM ION, SODIUM ION, ... | | 著者 | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQR
 
 | | Sialidase BT_1020 | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-L-arabinobiosidase, ... | | 著者 | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-22 | | 最終更新日 | 2017-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
