2N9B
 
 | |
2CPP
 
 | |
4RAY
 
 | |
4RAZ
 
 | |
4RB3
 
 | |
2HK6
 
 | |
4NKN
 
 | |
3FM0
 
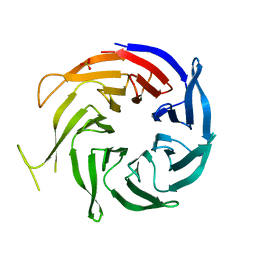 | | Crystal structure of WD40 protein Ciao1 | | 分子名称: | Protein CIAO1, SULFATE ION | | 著者 | Dong, A, Ravichandran, M, Crombet, L, Cossar, D, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | 登録日 | 2008-12-19 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
4O8U
 
 | | Structure of PF2046 | | 分子名称: | Uncharacterized protein PF2046 | | 著者 | Su, J, Liu, Z.-J. | | 登録日 | 2013-12-30 | | 公開日 | 2014-04-30 | | 実験手法 | X-RAY DIFFRACTION (2.345 Å) | | 主引用文献 | Crystal structure of a novel non-Pfam protein PF2046 solved using low resolution B-factor sharpening and multi-crystal averaging methods
Protein Cell, 1, 2010
|
|
2AK3
 
 | |
2XJ4
 
 | |
4O97
 
 | | Crystal structure of matriptase in complex with inhibitor | | 分子名称: | N-(trans-4-aminocyclohexyl)-3,5-bis[(3-carbamimidoylbenzyl)oxy]benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | 著者 | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | 登録日 | 2014-01-02 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4O9V
 
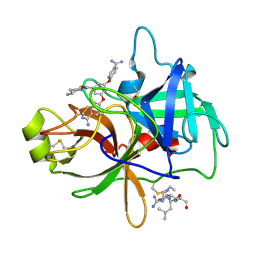 | | Crystal structure of matriptase in complex with inhibitor | | 分子名称: | N-(trans-4-aminocyclohexyl)-3,5-bis(4-carbamimidoylphenoxy)benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | 著者 | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | 登録日 | 2014-01-03 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4OE9
 
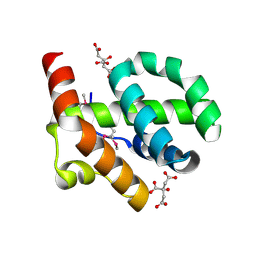 | |
4PUB
 
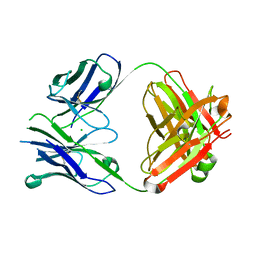 | | Crystal structure of Fab DX-2930 | | 分子名称: | CHLORIDE ION, DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN | | 著者 | Abendroth, J, Edwards, T.E, Nixon, A, Ladner, R. | | 登録日 | 2014-03-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
2ISC
 
 | | Crystal structure of Purine Nucleoside Phosphorylase from Trichomonas vaginalis with DADMe-Imm-A | | 分子名称: | (3R,4R)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-(HYDROXYMETHYL)PYRROLIDIN-3-OL, PHOSPHATE ION, purine nucleoside phosphorylase | | 著者 | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | 登録日 | 2006-10-17 | | 公開日 | 2007-06-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Inhibition and structure of Trichomonas vaginalis purine nucleoside phosphorylase with picomolar transition state analogues
Biochemistry, 46, 2007
|
|
1YBI
 
 | | Crystal structure of HA33A, a neurotoxin-associated protein from Clostridium botulinum type A | | 分子名称: | non-toxin haemagglutinin HA34 | | 著者 | Arndt, J.W, Gu, J, Jaroszewski, L, Schwarzenbacher, R, Hanson, M, Lebeda, F.J, Stevens, R.C. | | 登録日 | 2004-12-20 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Structure of the Neurotoxin-associated Protein HA33/A from Clostridium botulinum Suggests a Reoccurring beta-Trefoil Fold in the Progenitor Toxin Complex.
J.Mol.Biol., 346, 2005
|
|
7JTZ
 
 | | Yeast Glo3 GAP domain | | 分子名称: | ADP-ribosylation factor GTPase-activating protein GLO3, GLYCEROL, ZINC ION | | 著者 | Xie, B, Jackson, L.P. | | 登録日 | 2020-08-18 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | The Glo3 GAP crystal structure supports the molecular niche model for ArfGAPs in COPI coats.
Adv Biol Regul, 79, 2021
|
|
4OGX
 
 | | Crystal structure of Fab DX-2930 in complex with human plasma kallikrein at 2.4 Angstrom resolution | | 分子名称: | DX-2930 HEAVY CHAIN, DX-2930 LIGHT CHAIN, Plasma kallikrein, ... | | 著者 | Edwards, T.E, Clifton, M.C, Abendroth, J, Nixon, A, Ladner, R. | | 登録日 | 2014-01-16 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Inhibition of plasma kallikrein by a highly specific active site blocking antibody.
J.Biol.Chem., 289, 2014
|
|
4N14
 
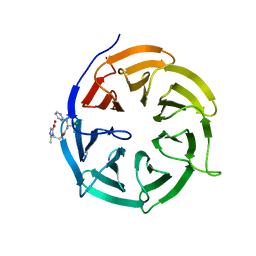 | | Crystal structure of Cdc20 and apcin complex | | 分子名称: | 2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl [(1R)-2,2,2-trichloro-1-(pyrimidin-2-ylamino)ethyl]carbamate, Cell division cycle protein 20 homolog | | 著者 | Luo, X, Tian, W, Yu, H. | | 登録日 | 2013-10-03 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C.
Nature, 514, 2014
|
|
2XJ9
 
 | |
2MY7
 
 | |
7K0R
 
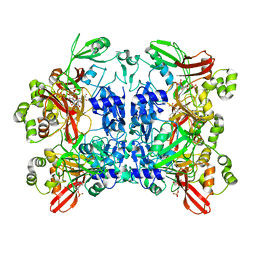 | | Nucleotide bound SARS-CoV-2 Nsp15 | | 分子名称: | PHOSPHATE ION, URIDINE-5'-MONOPHOSPHATE, Uridylate-specific endoribonuclease | | 著者 | Pillon, M.C, Stanley, R.E. | | 登録日 | 2020-09-04 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics.
Nat Commun, 12, 2021
|
|
4QDJ
 
 | |
4QDK
 
 | |
