6JWI
 
 | |
7C37
 
 | | Crystal structure of AofleA from Arthrobotrys oligospora | | 分子名称: | AofleA, BICINE | | 著者 | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | 登録日 | 2020-05-11 | | 公開日 | 2020-07-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7CTQ
 
 | | Peptidyl tryptophan dihydroxylase QhpG essential for tryptophylquinone cofactor biogenesis | | 分子名称: | (2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-6-(cyclohexylmethoxy)-2-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol, FLAVIN-ADENINE DINUCLEOTIDE, HEXANE-1,6-DIOL, ... | | 著者 | Oozeki, T, Nakai, T, Okajima, T. | | 登録日 | 2020-08-20 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.978 Å) | | 主引用文献 | Functional and structural characterization of a flavoprotein monooxygenase essential for biogenesis of tryptophylquinone cofactor.
Nat Commun, 12, 2021
|
|
6JWH
 
 | | Yeast Npl4 zinc finger, MPN and CTD domains | | 分子名称: | GLYCEROL, Nuclear protein localization protein 4, ZINC ION | | 著者 | Sato, Y, Fukai, S. | | 登録日 | 2019-04-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.72000253 Å) | | 主引用文献 | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
7D56
 
 | |
7D5V
 
 | | Structure of the C646A mutant of peptidylarginine deiminase type III (PAD3) | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Protein-arginine deiminase type-3 | | 著者 | Akimoto, M, Mashimo, R, Unno, M. | | 登録日 | 2020-09-28 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
7DAN
 
 | |
7D5R
 
 | | Structure of the Ca2+-bound C646A mutant of peptidylarginine deiminase type III (PAD3) | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Mashimo, R, Akimoto, M, Unno, M. | | 登録日 | 2020-09-28 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.148 Å) | | 主引用文献 | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
7D4Y
 
 | |
7D8N
 
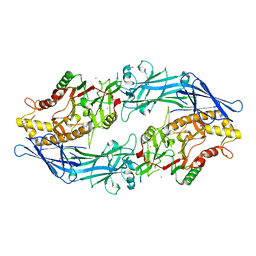 | | Structure of the inactive form of wild-type peptidylarginine deiminase type III (PAD3) crystallized under the condition with high concentrations of Ca2+ | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Funabashi, K, Sawata, M, Unno, M. | | 登録日 | 2020-10-08 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.753 Å) | | 主引用文献 | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
6I7O
 
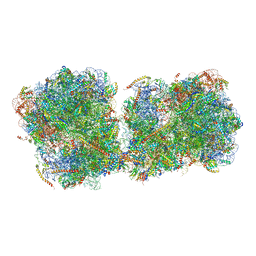 | | The structure of a di-ribosome (disome) as a unit for RQC and NGD quality control pathways recognition. | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Tesina, P, Cheng, J, Becker, T, Beckmann, R. | | 登録日 | 2018-11-16 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways.
EMBO J., 38, 2019
|
|
7DE9
 
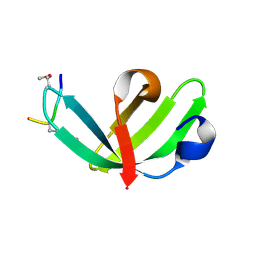 | |
6JWJ
 
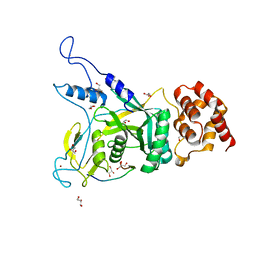 | | Npl4 in complex with Ufd1 | | 分子名称: | GLYCEROL, Nuclear protein localization protein 4, Peptide from Ubiquitin fusion degradation protein 1, ... | | 著者 | Sato, Y, Fukai, S. | | 登録日 | 2019-04-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
376D
 
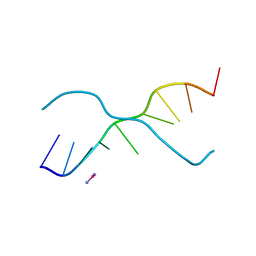 | | A ZIPPER-LIKE DNA DUPLEX D(GCGAAAGCT) | | 分子名称: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*(CBR)P*GP*AP*AP*AP*GP*CP*T)-3') | | 著者 | Cruse, W.B.T, Shepard, W, Prange, T, delalFortelle, E, Fourme, R. | | 登録日 | 1998-01-22 | | 公開日 | 1999-10-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A zipper-like duplex in DNA: the crystal structure of d(GCGAAAGCT) at 2.1 A resolution.
Structure, 6, 1998
|
|
1O5R
 
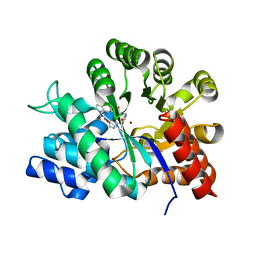 | | Crystal structure of adenosine deaminase complexed with a potent inhibitor | | 分子名称: | 1-[(1R)-3-(6-{[(BENZYLAMINO)CARBONYL]AMINO}-1H-INDOL-1-YL)-1-(HYDROXYMETHYL)PROPYL]-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | 著者 | Kinoshita, T. | | 登録日 | 2003-10-05 | | 公開日 | 2004-09-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure-based design, synthesis, and structure-activity relationship studies of novel non-nucleoside adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
1OD6
 
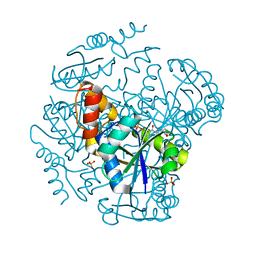 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | 分子名称: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | 著者 | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | 登録日 | 2003-02-13 | | 公開日 | 2003-03-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7JMA
 
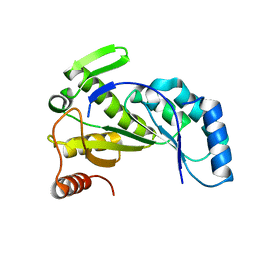 | |
3VUQ
 
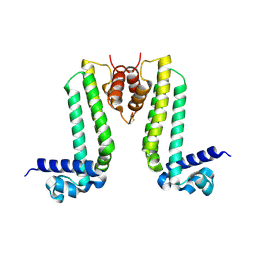 | | Crystal structure of TTHA0167, a transcriptional regulator, TetR/AcrR family from Thermus thermophilus HB8 | | 分子名称: | Transcriptional regulator (TetR/AcrR family) | | 著者 | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | 登録日 | 2012-07-04 | | 公開日 | 2013-02-27 | | 最終更新日 | 2013-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure and function of a TetR family transcriptional regulator, SbtR, from thermus thermophilus HB8
Proteins, 81, 2013
|
|
7JMB
 
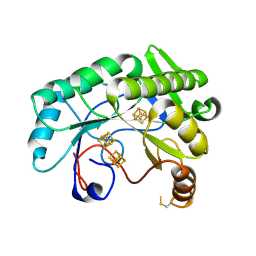 | | Crystal structure of Nitrogenase iron-molybdenum cofactor biosynthesis enzyme NifB from Methanothermobacter thermautotrophicus with three Fe4S4 clusters | | 分子名称: | IRON/SULFUR CLUSTER, Nitrogenase iron-molybdenum cofactor biosynthesis protein NifB | | 著者 | Kang, W, Rettberg, L, Ribbe, M.W, Hu, Y. | | 登録日 | 2020-07-31 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-Ray Crystallographic Analysis of NifB with a Full Complement of Clusters: Structural Insights into the Radical SAM-Dependent Carbide Insertion During Nitrogenase Cofactor Assembly.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3AHO
 
 | | PZ PEPTIDASE A with inhibitor 2 | | 分子名称: | 1-{3-[(R)-{(1R)-1-[(glycyl-L-prolyl)amino]-2-phenylethyl}(hydroxy)phosphoryl]propanoyl}-L-prolyl-D-norleucine, ACETATE ION, Oligopeptidase, ... | | 著者 | Nakano, H. | | 登録日 | 2010-04-25 | | 公開日 | 2010-09-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The exquisite structure and reaction mechanism of bacterial Pz-peptidase A toward collagenous peptides: X-ray crystallographic structure analysis of PZ-peptidase a reveals differences from mammalian thimet oligopeptidase.
J.Biol.Chem., 285, 2010
|
|
8OOR
 
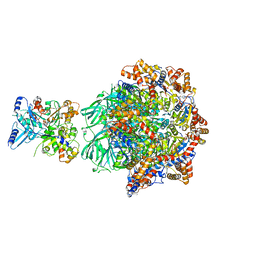 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOK
 
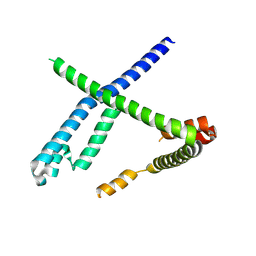 | |
8OOF
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO7
 
 | | CryoEM Structure INO80core Hexasome complex composite model state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
