3C8A
 
 | |
3U5R
 
 | |
2AW9
 
 | |
1L1P
 
 | | Solution Structure of the PPIase Domain from E. coli Trigger Factor | | 分子名称: | trigger factor | | 著者 | Kozlov, G, Trempe, J.-F, Perreault, A, Wong, M, Denisov, A, Ghandi, S, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2002-02-19 | | 公開日 | 2003-06-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Closed Form of a Peptidyl-Prolyl Isomerase Reveals the Mechanism of Protein Folding
To be Published
|
|
4RIS
 
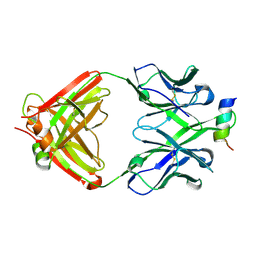 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | 分子名称: | CH58-UA Fab heavy chain, CH58-UA Fab light chain, Envelope glycoprotein | | 著者 | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | 登録日 | 2014-10-07 | | 公開日 | 2015-08-12 | | 最終更新日 | 2015-09-02 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
4RIR
 
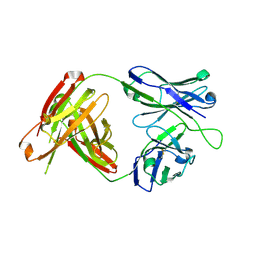 | | Structural Analysis of the Unmutated Ancestor of the HIV-1 Envelope V2 Region Antibody CH58 Isolated From an RV144 HIV-1 Vaccine Efficacy Trial Vaccinee and Associated with Decreased Transmission Risk | | 分子名称: | CH58-UA Fab heavy chain, CH58-UA Fab light chain | | 著者 | Nicely, N.I, Wiehe, K, Kepler, T.B, Jaeger, F.H, Dennison, S.M, Liao, H.-X, Alam, S.M, Hwang, K.-K, Bonsignori, M, Rerks-Ngarm, S, Nitayaphan, S, Pitisuttithum, P, Kaewkungwal, J, Robb, M.L, O'Connell, R.J, Michael, N.L, Kim, J.H, Haynes, B.F. | | 登録日 | 2014-10-07 | | 公開日 | 2015-08-12 | | 最終更新日 | 2015-09-02 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee.
EBioMedicine, 2, 2015
|
|
7JSN
 
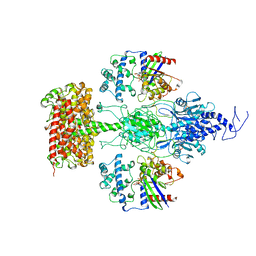 | | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6 | | 分子名称: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Gao, Y, Eskici, G, Ramachandran, S, Skiniotis, G, Cerione, R.A. | | 登録日 | 2020-08-15 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6.
Mol.Cell, 80, 2020
|
|
3BQT
 
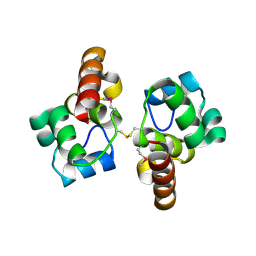 | |
3BZW
 
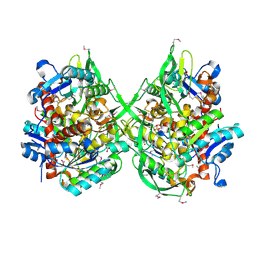 | | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron | | 分子名称: | ACETATE ION, Putative lipase, SULFATE ION | | 著者 | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-01-18 | | 公開日 | 2008-02-05 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron.
To be Published
|
|
3S4P
 
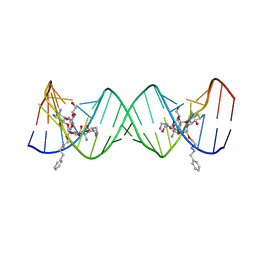 | | Crystal structure of the bacterial ribosomal decoding site complexed with an amphiphilic paromomycin O2''-ether analogue | | 分子名称: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-2-O-{2-[(2-phenylethyl)amino]ethyl}-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | 著者 | Szychowski, J, Kondo, J, Zahr, O, Auclair, K, Westhof, E, Hanessian, S, Keillor, J.W. | | 登録日 | 2011-05-20 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Inhibition of aminoglycoside-deactivating enzymes APH(3')-IIIa and AAC(6')-Ii by amphiphilic paromomycin O2''-ether analogues
Chemmedchem, 6, 2011
|
|
3BWI
 
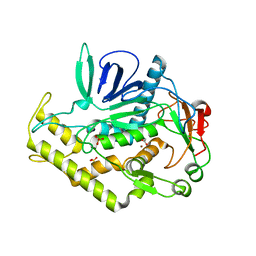 | | Crystal structure of the catalytic domain of botulinum neurotoxin serotype A with an acetate ion bound at the active site | | 分子名称: | ACETATE ION, Botulinum neurotoxin A light chain, SULFATE ION, ... | | 著者 | Kumaran, D, Rawat, R, Swaminathan, S. | | 登録日 | 2008-01-09 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure- and Substrate-based Inhibitor Design for Clostridium botulinum Neurotoxin Serotype A
J.Biol.Chem., 283, 2008
|
|
3T2M
 
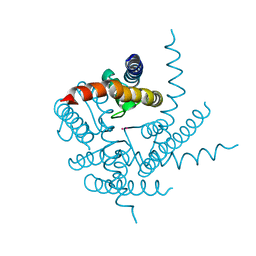 | | Crystal Structure of NaK Channel N68D Mutant | | 分子名称: | POTASSIUM ION, Potassium channel protein | | 著者 | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | 登録日 | 2011-07-22 | | 公開日 | 2011-10-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.953 Å) | | 主引用文献 | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2G6T
 
 | |
3T1C
 
 | | Crystal Structure of NaK Channel D66Y Mutant | | 分子名称: | POTASSIUM ION, Potassium channel protein | | 著者 | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | 登録日 | 2011-07-21 | | 公開日 | 2011-10-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3LMU
 
 | | Crystal structure of DTD from Plasmodium falciparum | | 分子名称: | D-tyrosyl-tRNA(Tyr) deacylase, IODIDE ION | | 著者 | Manickam, Y, Bhatt, T.K, Khan, S, Sharma, A. | | 登録日 | 2010-02-01 | | 公開日 | 2010-03-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4JFC
 
 | | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666 | | 分子名称: | Enoyl-CoA hydratase, GLYCEROL | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-02-28 | | 公開日 | 2013-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of a enoyl-CoA hydratase from Polaromonas sp. JS666
To be Published
|
|
2F4N
 
 | |
3WBD
 
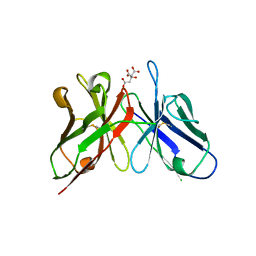 | | Crystal structure of anti-polysialic acid antibody single chain Fv fragment (mAb735) complexed with octasialic acid | | 分子名称: | CITRATE ANION, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid, single chain Fv fragment of mAb735 | | 著者 | Nagae, M, Ikeda, A, Hanashima, S, Kitajima, K, Sato, C, Yamaguchi, Y. | | 登録日 | 2013-05-14 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of anti-polysialic acid antibody single chain Fv fragment complexed with octasialic acid: insight into the binding preference for polysialic acid.
J.Biol.Chem., 288, 2013
|
|
4OO9
 
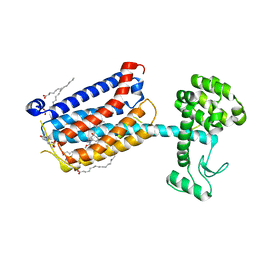 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator mavoglurant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mavoglurant, Metabotropic glutamate receptor 5, ... | | 著者 | Dore, A.S, Okrasa, K, Patel, J.C, Serrano-Vega, M, Bennett, K, Cooke, R.M, Errey, J.C, Jazayeri, A, Khan, S, Tehan, B, Weir, M, Wiggin, G.R, Marshall, F.H. | | 登録日 | 2014-01-31 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain.
Nature, 511, 2014
|
|
3C8B
 
 | |
3C89
 
 | |
4JCU
 
 | | Crystal structure of a 5-carboxymethyl-2-hydroxymuconate isomerase from Deinococcus radiodurans R1 | | 分子名称: | 5-carboxymethyl-2-hydroxymuconate isomerase | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-02-22 | | 公開日 | 2013-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a 5-carboxymethyl-2-hydroxymuconate isomerase from Deinococcus radiodurans R1
To be Published
|
|
3C88
 
 | |
2JML
 
 | | Solution structure of the N-terminal domain of CarA repressor | | 分子名称: | DNA BINDING DOMAIN/TRANSCRIPTIONAL REGULATOR | | 著者 | Jimenez, M, Padmanabhan, S, Gonzalez, C, Perez-Marin, M.C, Elias-Arnanz, M, Murillo, F.J, Rico, M. | | 登録日 | 2006-11-20 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for operator and antirepressor recognition by Myxococcus xanthus CarA repressor.
Mol.Microbiol., 63, 2007
|
|
3BT3
 
 | |
