6LQH
 
 | | High resolution architecture of curli complex | | 分子名称: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | 著者 | Zhang, M, Shi, H, Huang, Y. | | 登録日 | 2020-01-13 | | 公開日 | 2020-07-15 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
6LQJ
 
 | | Low resolution architecture of curli complex | | 分子名称: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | 著者 | Zhang, M, Shi, H, Huang, Y. | | 登録日 | 2020-01-13 | | 公開日 | 2020-07-15 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
6M67
 
 | | The Cryo-EM Structure of Human Pannexin 1 with D376E/D379E Mutation | | 分子名称: | Pannexin-1 | | 著者 | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | 登録日 | 2020-03-13 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
6LQN
 
 | | EBV tegument protein BBRF2 | | 分子名称: | Cytoplasmic envelopment protein 1, NITRATE ION, SULFATE ION | | 著者 | He, H.P, Luo, M, Cao, Y.L, Gao, S. | | 登録日 | 2020-01-14 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.600022 Å) | | 主引用文献 | Structure of Epstein-Barr virus tegument protein complex BBRF2-BSRF1 reveals its potential role in viral envelopment.
Nat Commun, 11, 2020
|
|
7P0A
 
 | |
7P0T
 
 | |
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | 著者 | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | 登録日 | 2022-02-21 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
7FAP
 
 | | Structure of VAR2CSA-CSA 3D7 | | 分子名称: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Erythrocyte membrane protein 1, PfEMP1 | | 著者 | Wang, L, Wang, Z. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
8I3V
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant escitalopram in an inward-open state at resolution of 2.85 angstrom. | | 分子名称: | (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | 著者 | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | 登録日 | 2023-01-18 | | 公開日 | 2024-07-03 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
5ULA
 
 | |
8ZET
 
 | | Tp-PSI-FCPI-S in Thalassiosira pseudonana | | 分子名称: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | 著者 | Feng, Y, Li, Z, Shen, J.R, Wang, W. | | 登録日 | 2024-05-06 | | 公開日 | 2024-12-25 | | 最終更新日 | 2025-04-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of PSI-FCPI from Thalassiosira pseudonana grown under high light provide evidence for convergent evolution and light-adaptive strategies in diatom FCPIs.
J Integr Plant Biol, 67, 2025
|
|
8ZEH
 
 | | PSI-FCPI-L in Thalassiosira pseudonana | | 分子名称: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | 著者 | Feng, Y, Li, Z, Wang, W, Shen, J.R. | | 登録日 | 2024-05-06 | | 公開日 | 2024-12-25 | | 最終更新日 | 2025-04-30 | | 実験手法 | ELECTRON MICROSCOPY (2.78 Å) | | 主引用文献 | Structures of PSI-FCPI from Thalassiosira pseudonana grown under high light provide evidence for convergent evolution and light-adaptive strategies in diatom FCPIs.
J Integr Plant Biol, 67, 2025
|
|
7ESI
 
 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 at 1. 8 angstrom resolution. | | 分子名称: | CALCIUM ION, Collagenase unit (CU), Peptide P1, ... | | 著者 | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | 登録日 | 2021-05-11 | | 公開日 | 2022-02-09 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun, 13, 2022
|
|
4UAB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Chromohalobacter salexigens DSM 3043 (Csal_0678), Target EFI-501078, with bound ethanolamine | | 分子名称: | CHLORIDE ION, ETHANOLAMINE, Twin-arginine translocation pathway signal | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-08-08 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4W93
 
 | | Human pancreatic alpha-amylase in complex with montbretin A | | 分子名称: | CALCIUM ION, CHLORIDE ION, Montbretin A, ... | | 著者 | Williams, L.K, Caner, S, Brayer, G.D. | | 登録日 | 2014-08-27 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.352 Å) | | 主引用文献 | The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif.
Nat.Chem.Biol., 11, 2015
|
|
6GB5
 
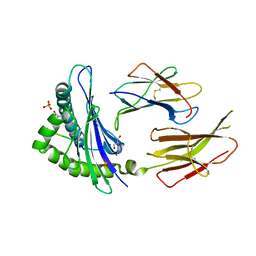 | | Structure of H-2Db with truncated SEV peptide and GL | | 分子名称: | Beta-2-microglobulin, GLY-LEU, GLYCEROL, ... | | 著者 | Hafstrand, I, Sandalova, T, Achour, A. | | 登録日 | 2018-04-13 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GB7
 
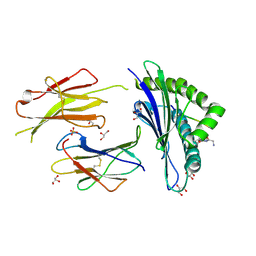 | | Structure of H-2Db with scoop loop from tapasin | | 分子名称: | Beta-2-microglobulin, GLY-GLY-LEU-SER, GLYCEROL, ... | | 著者 | Hafstrand, I, Sandalova, T, Achour, A. | | 登録日 | 2018-04-13 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GB6
 
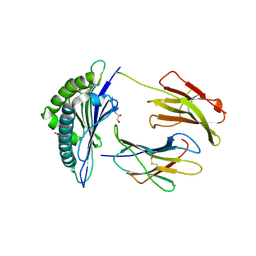 | | Structure of H-2Kb with dipeptide GL | | 分子名称: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | 著者 | Hafstrand, I, Sandalova, T, Achour, A. | | 登録日 | 2018-04-13 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8IW9
 
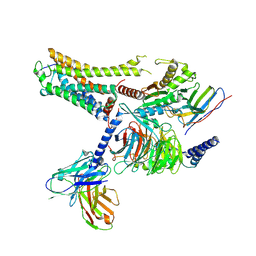 | | Cryo-EM structure of the CAD-bound mTAAR9-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW4
 
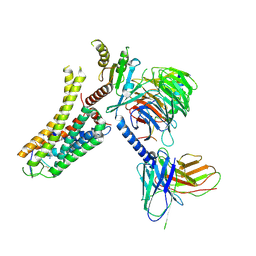 | | Cryo-EM structure of the SPE-bound mTAAR9-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW7
 
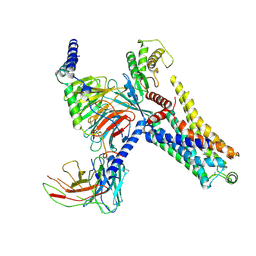 | | Cryo-EM structure of the PEA-bound mTAAR9-Gs complex | | 分子名称: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8ITF
 
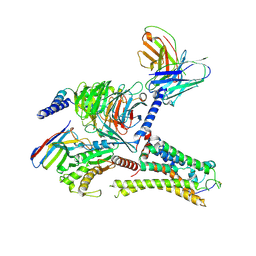 | | Cryo-EM structure of the DMCHA-bound mTAAR9-Gs complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-22 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWE
 
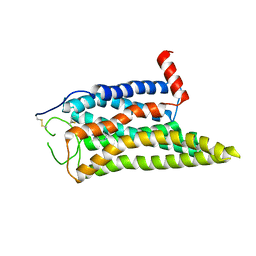 | | Cryo-EM structure of the SPE-mTAAR9 complex | | 分子名称: | SPERMIDINE, Trace amine-associated receptor 9 | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IWM
 
 | | Cryo-EM structure of the PEA-bound mTAAR9 complex | | 分子名称: | 2-PHENYLETHYLAMINE, Trace amine-associated receptor 9 | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-30 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
8IW1
 
 | | Cryo-EM structure of the PEA-bound mTAAR9-Golf complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(olf) subunit alpha, ... | | 著者 | Sun, J.P, Li, Q, Yang, F, Xu, Y.F, Guo, L.L, Lian, S, Zhang, M.H, Rong, N.K. | | 登録日 | 2023-03-29 | | 公開日 | 2023-05-31 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of amine odorant perception by a mammal olfactory receptor.
Nature, 618, 2023
|
|
