3G02
 
 | |
6SGS
 
 | | Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc | | 分子名称: | DARPin, Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrB | | 著者 | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | 登録日 | 2019-08-05 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
8WOV
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, G233A mutant | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | 著者 | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | 登録日 | 2023-10-07 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
5T6L
 
 | |
1S2X
 
 | | Crystal structure of Cag-Z from Helicobacter pylori | | 分子名称: | Cag-Z, ISOPROPYL ALCOHOL | | 著者 | Cendron, L, Seydel, A, Angelini, A, Battistutta, R, Zanotti, G. | | 登録日 | 2004-01-12 | | 公開日 | 2004-07-27 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of CagZ, a protein from the Helicobacter pylori pathogenicity island that encodes for a type IV secretion system
J.Mol.Biol., 340, 2004
|
|
1S1N
 
 | | SH3 domain of human nephrocystin | | 分子名称: | Nephrocystin 1 | | 著者 | Le Maire, A, Weber, T, Saunier, S, Antignac, C, Ducruix, A, Dardel, F. | | 登録日 | 2004-01-07 | | 公開日 | 2005-01-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the SH3 domain of human nephrocystin and analysis of a mutation-causing juvenile nephronophthisis.
Proteins, 59, 2005
|
|
4EN6
 
 | | Crystal structure of HA70 (HA3) subcomponent of Clostridium botulinum type C progenitor toxin in complex with alpha 2-3-sialyllactose | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, Hemagglutinin components HA-22/23/53, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Yamashita, S, Yoshida, H, Tonozuka, T, Nishikawa, A, Kamitori, S. | | 登録日 | 2012-04-12 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Carbohydrate recognition mechanism of HA70 from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides
Febs Lett., 586, 2012
|
|
6SUA
 
 | |
5NJ1
 
 | |
5T35
 
 | | The PROTAC MZ1 in complex with the second bromodomain of Brd4 and pVHL:ElonginC:ElonginB | | 分子名称: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Transcription elongation factor B polypeptide 1, ... | | 著者 | Gadd, M.S, Zengerle, M, Ciulli, A. | | 登録日 | 2016-08-24 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of PROTAC cooperative recognition for selective protein degradation.
Nat. Chem. Biol., 13, 2017
|
|
8RXR
 
 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | 分子名称: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | 著者 | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | 登録日 | 2024-02-07 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 2024
|
|
5AFL
 
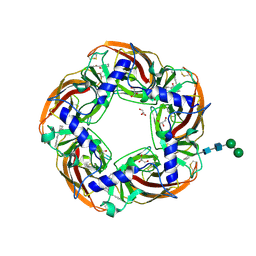 | | alpha7-AChBP in complex with lobeline and fragment 3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE-BINDING PROTEIN, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-7, ... | | 著者 | Spurny, R, Debaveye, S, Farinha, A, Veys, K, Gossas, T, Atack, J, Bertrand, D, Kemp, J, Vos, A, Danielson, U.H, Tresadern, G, Ulens, C. | | 登録日 | 2015-01-22 | | 公開日 | 2015-05-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.385 Å) | | 主引用文献 | Molecular Blueprint of Allosteric Binding Sites in a Homologue of the Agonist-Binding Domain of the Alpha7 Nicotinic Acetylcholine Receptor.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8R88
 
 | |
5M1T
 
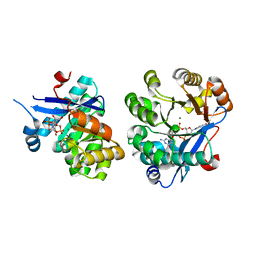 | | PaMucR Phosphodiesterase, c-di-GMP complex | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, MucR Phosphodiesterase | | 著者 | Hutchin, A, Tews, I, Walsh, M.A. | | 登録日 | 2016-10-10 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
3S8Y
 
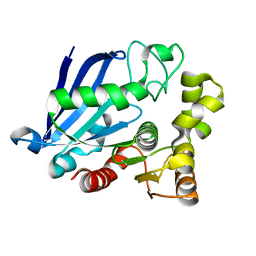 | | Bromide soaked structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | 分子名称: | BROMIDE ION, Esterase APC40077 | | 著者 | Petit, P, Dong, A, Kagan, O, Savchenko, A, Yakunin, A.F. | | 登録日 | 2011-05-31 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
2IGS
 
 | | Crystal Structure of the Protein of Unknown Function from Pseudomonas aeruginosa | | 分子名称: | ACETIC ACID, GLYCEROL, Hypothetical protein, ... | | 著者 | Kim, Y, Joachimiak, A, Skarina, T, Egorova, O, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-09-25 | | 公開日 | 2006-10-24 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Crystal Structure of the Hypothetical Protein from Pseudomonas aeruginosa
To be Published
|
|
5M4K
 
 | | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase | | 分子名称: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, ... | | 著者 | Baker, L.M, Brough, P, Surgenor, A. | | 登録日 | 2016-10-18 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Application of Off-Rate Screening in the Identification of Novel Pan-Isoform Inhibitors of Pyruvate Dehydrogenase Kinase.
J. Med. Chem., 60, 2017
|
|
5A7N
 
 | | Crystal structure of human JMJD2A in complex with compound 43 | | 分子名称: | 1,2-ETHANEDIOL, 2-(5-cyano-2-oxidanyl-phenyl)pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | 著者 | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Williams, E, Riesebos, E, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | 登録日 | 2015-07-09 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5LSK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C COMPLEX | | 分子名称: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | 著者 | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | 登録日 | 2016-09-02 | | 公開日 | 2016-11-16 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.502 Å) | | 主引用文献 | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
3OH3
 
 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE -Arabinose | | 分子名称: | GLYCEROL, UDP-sugar pyrophosphorylase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2S,3R,4S,5S)-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | 著者 | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | 登録日 | 2010-08-17 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
5LUX
 
 | | Homeobox transcription factor CDX1 bound to methylated DNA | | 分子名称: | DNA (5'-D(P*GP*AP*GP*GP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*CP*CP*TP*C)-3'), ... | | 著者 | Morgunova, E, Popov, A, Taipale, J. | | 登録日 | 2016-09-12 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.23 Å) | | 主引用文献 | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
194L
 
 | | THE 1.40 A STRUCTURE OF SPACEHAB-01 HEN EGG WHITE LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Vaney, M.C, Maignan, S, Ries-Kautt, M, Ducruix, A. | | 登録日 | 1995-09-01 | | 公開日 | 1995-12-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | High-resolution structure (1.33 A) of a HEW lysozyme tetragonal crystal grown in the APCF apparatus. Data and structural comparison with a crystal grown under microgravity from SpaceHab-01 mission.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6SL7
 
 | |
5SY8
 
 | |
2IDA
 
 | | Solution NMR Structure of Protein RPA1320 from Rhodopseudomonas Palustris. Northeast Structural Genomics Consortium Target RpT3; Ontario Center for Structural Proteomics Target RP1313. | | 分子名称: | Hypothetical protein, ZINC ION | | 著者 | Lemak, A, Yee, A, Lukin, J.A, Karra, M, Gutmanas, A, Guido, V, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-09-14 | | 公開日 | 2006-10-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of RPA1320
To be Published
|
|
