7UXR
 
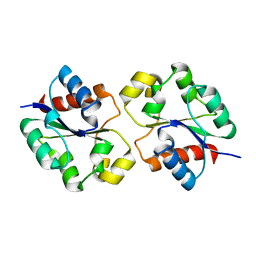 | | Crystal structure of the BtTir TIR domain | | 分子名称: | TIR domain protein | | 著者 | Shi, Y, Masic, V, Mosaiab, T, Vasquez, E, Ve, T. | | 登録日 | 2022-05-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXU
 
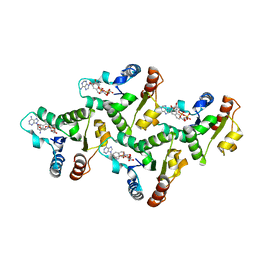 | | CryoEM structure of the TIR domain from AbTir in complex with 3AD | | 分子名称: | Molecular chaperone Tir, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Li, S, Nanson, J.D, Manik, M.K, Gu, W, Landsberg, M.J, Ve, T, Kobe, B. | | 登録日 | 2022-05-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXS
 
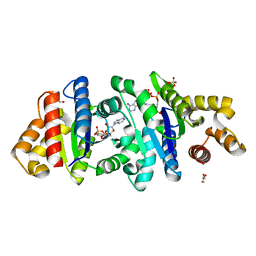 | | Crystal structure of the BcThsA SLOG domain in complex with 3'cADPR | | 分子名称: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, BcThsA, GLYCEROL, ... | | 著者 | Shi, Y, Masic, V, Mosaiab, T, Ve, T. | | 登録日 | 2022-05-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXT
 
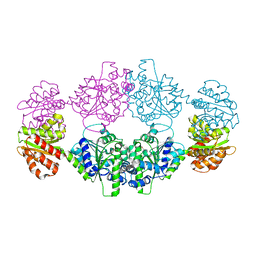 | | Crystal structure of ligand-free SeThsA | | 分子名称: | GLYCEROL, TRIETHYLENE GLYCOL, USG protein | | 著者 | Shi, Y, Masic, V, Mosaiab, T, Nanson, J.D, Kobe, B, Ve, T. | | 登録日 | 2022-05-06 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
3DCV
 
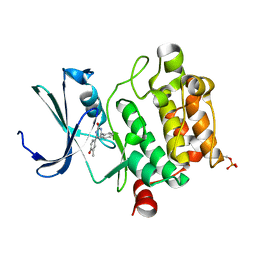 | | Crystal structure of human Pim1 kinase complexed with 4-(4-hydroxy-3-methyl-phenyl)-6-phenylpyrimidin-2(1H)-one | | 分子名称: | 4-(4-hydroxy-3-methylphenyl)-6-phenylpyrimidin-2(5H)-one, Proto-oncogene serine/threonine-protein kinase Pim-1 | | 著者 | Bellamacina, C.R, Shafer, C.M, Lindvall, M, Gesner, T.G, Yabannavar, A, Weiping, J, Song, L, Walter, A. | | 登録日 | 2008-06-04 | | 公開日 | 2008-08-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | 4-(1H-indazol-5-yl)-6-phenylpyrimidin-2(1H)-one analogs as potent CDC7 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7E7Y
 
 | |
7E7X
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E88
 
 | |
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
7BYR
 
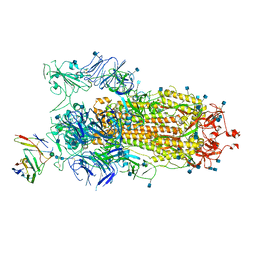 | | BD23-Fab in complex with the S ectodomain trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab23-Fab-Heavy Chain, ... | | 著者 | Zhu, Q, Wang, G, Xiao, J. | | 登録日 | 2020-04-24 | | 公開日 | 2020-06-10 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells.
Cell, 182, 2020
|
|
5YZ0
 
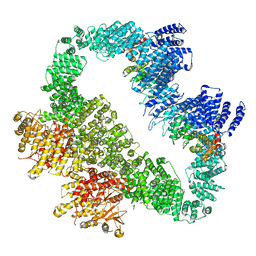 | | Cryo-EM Structure of human ATR-ATRIP complex | | 分子名称: | ATR-interacting protein, Serine/threonine-protein kinase ATR | | 著者 | Rao, Q, Liu, M, Tian, Y, Wu, Z, Wang, H, Wang, J, Xu, Y. | | 登録日 | 2017-12-11 | | 公開日 | 2018-01-31 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Cryo-EM structure of human ATR-ATRIP complex.
Cell Res., 28, 2018
|
|
6MF8
 
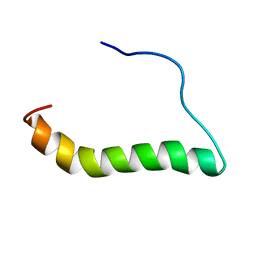 | | TCR alpha transmembrane domain | | 分子名称: | T-cell receptor alpha chain C region | | 著者 | Brazin, K.N, Reinherz, E.L. | | 登録日 | 2018-09-10 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The T Cell Antigen Receptor alpha Transmembrane Domain Coordinates Triggering through Regulation of Bilayer Immersion and CD3 Subunit Associations.
Immunity, 49, 2018
|
|
7YC2
 
 | |
7XYT
 
 | |
7XV7
 
 | |
7XYV
 
 | |
7XYW
 
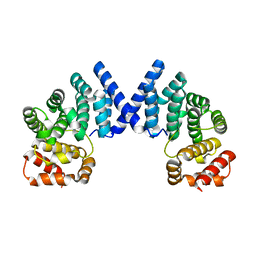 | |
7XYX
 
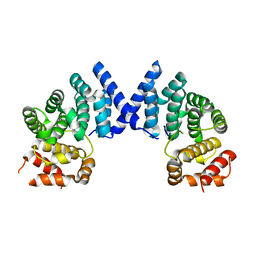 | |
7XYS
 
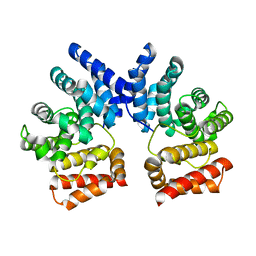 | |
7XYU
 
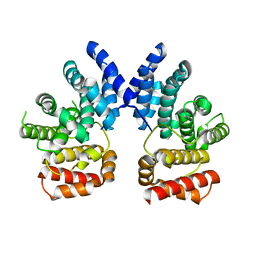 | |
7W3S
 
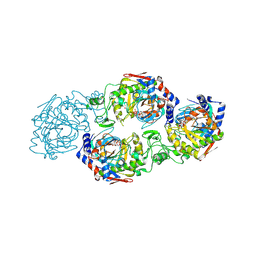 | | The complex structure of Larg1-ADPr from Legionella pneumophila | | 分子名称: | Type IV secretion protein Dot, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Ouyang, S, Guan, H, Li, P. | | 登録日 | 2021-11-26 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-05-18 | | 実験手法 | X-RAY DIFFRACTION (2.324 Å) | | 主引用文献 | Legionella pneumophila temporally regulates the activity of ADP/ATP translocases by reversible ADP-ribosylation.
mLife, 2022
|
|
7XQL
 
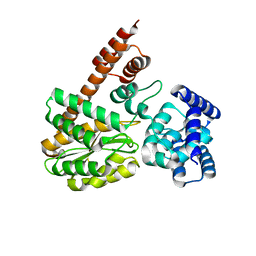 | | complex structure of LegA15 with GNP | | 分子名称: | Ankyrin repeat-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Chen, T.T, Lin, Y.L. | | 登録日 | 2022-05-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.272 Å) | | 主引用文献 | Atypical Legionella GTPase effector hijacks host vesicular transport factor p115 to regulate host lipid droplet.
Sci Adv, 8, 2022
|
|
7Y3A
 
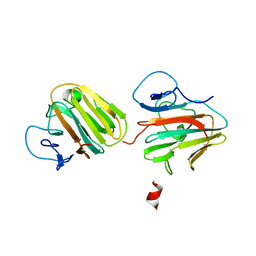 | | Crystal structure of TRIM7 bound to 2C | | 分子名称: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-2C | | 著者 | Dong, C, Yan, X. | | 登録日 | 2022-06-10 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
