4LCJ
 
 | | CtBP2 in complex with substrate MTOB | | 分子名称: | 4-(METHYLSULFANYL)-2-OXOBUTANOIC ACID, C-terminal-binding protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hilbert, B.J, Schiffer, C.A, Royer Jr, W.E. | | 登録日 | 2013-06-21 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Crystal structures of human CtBP in complex with substrate MTOB reveal active site features useful for inhibitor design.
Febs Lett., 588, 2014
|
|
4U6S
 
 | | CtBP1 in complex with substrate phenylpyruvate | | 分子名称: | 3-PHENYLPYRUVIC ACID, C-terminal-binding protein 1, CALCIUM ION, ... | | 著者 | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | 登録日 | 2014-07-29 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
4U6Q
 
 | | CtBP1 bound to inhibitor 2-(hydroxyimino)-3-phenylpropanoic acid | | 分子名称: | (2E)-2-(hydroxyimino)-3-phenylpropanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | 著者 | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | 登録日 | 2014-07-29 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
8FL5
 
 | |
9N8P
 
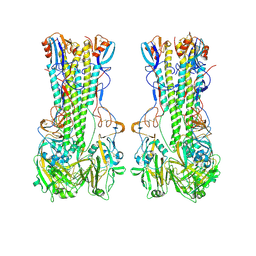 | | Subtomogram average of dimers of influenza HA trimers | | 分子名称: | Hemagglutinin, Hemagglutinin HA2 chain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose | | 著者 | Huang, Q.J, Song, K, Schiffer, C.A, Somasundaran, M. | | 登録日 | 2025-02-09 | | 公開日 | 2025-04-16 | | 最終更新日 | 2025-04-30 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Virion-associated influenza hemagglutinin clusters upon sialic acid binding visualized by cryoelectron tomography.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ELV
 
 | | Crystal Structure of SARS-CoV-2 Mpro mutant E166V with Pfizer Intravenous Inhibitor PF-00835231 | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | 著者 | Zvornicanin, S.N, Shaqra, A.M, Schiffer, C.A. | | 登録日 | 2024-12-05 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-05-21 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Molecular mechanisms of drug resistance and compensation in SARS-CoV-2 main protease: the interplay between E166 and L50.
Mbio, 16, 2025
|
|
2I0D
 
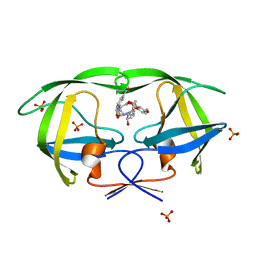 | | Crystal structure of AD-81 complexed with wild type HIV-1 protease | | 分子名称: | (5S)-3-(3-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, ACETATE ION, PHOSPHATE ION, ... | | 著者 | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | 登録日 | 2006-08-10 | | 公開日 | 2006-12-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
2I0A
 
 | | Crystal Structure of KB-19 complexed with wild type HIV-1 protease | | 分子名称: | (5S)-3-(4-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | 登録日 | 2006-08-10 | | 公開日 | 2006-12-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
5JQD
 
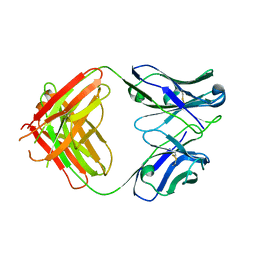 | | Antibody Fab Fragment | | 分子名称: | D80 Fab Fragment Heavy Chain, D80 Fab Fragment Light Chain | | 著者 | Zhang, Z, Prachanronarong, K, Gellatly, K, Marasco, W.A, Schiffer, C.A. | | 登録日 | 2016-05-04 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.591 Å) | | 主引用文献 | Structural Basis of an Influenza Hemagglutinin Stem-Directed Antibody Retaining the G6 Idiotype
To Be Published
|
|
5KEG
 
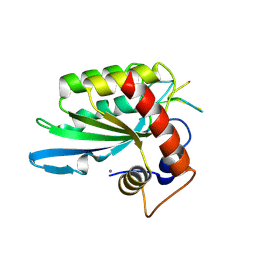 | | Crystal structure of APOBEC3A in complex with a single-stranded DNA | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*TP*TP*CP*TP*T)-3'), ... | | 著者 | Kouno, T, Hilbert, B.J, Silvas, T, Royer, W.E, Matsuo, H, Schiffer, C.A. | | 登録日 | 2016-06-09 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity.
Nat Commun, 8, 2017
|
|
5JO4
 
 | | Antibody Fab Fragment Complex | | 分子名称: | D80 Fab Heavy Chain, D80 Fab Light Chain, G6 Fab Heavy Chain, ... | | 著者 | Zhang, Z, Prachanronarong, K.P, Marasco, W.A, Schiffer, C.A.S. | | 登録日 | 2016-05-01 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Structural Basis of an Influenza Hemagglutinin Stem-Directed Antibody Retaining the G6 Idiotype
To Be Published
|
|
7MMD
 
 | |
8DSU
 
 | |
8E7N
 
 | | Crystal structure of beluga whale Gammacoronavirus SW1 Mpro with GC-376 captured in two conformational states | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, GLYCEROL, ... | | 著者 | Shaqra, A.M, Schiffer, C.A. | | 登録日 | 2022-08-24 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structures of Inhibitor-Bound Main Protease from Delta- and Gamma-Coronaviruses.
Viruses, 15, 2023
|
|
8E7T
 
 | |
8E5C
 
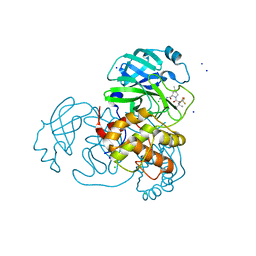 | | Crystal Structure of SARS CoV-2 Mpro mutant L50F with Nirmatrelvir captured in two conformational states | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | 著者 | Shaqra, A.M, Schiffer, C.A. | | 登録日 | 2022-08-20 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
8E4W
 
 | |
8DT9
 
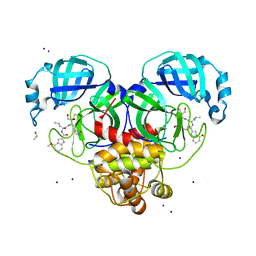 | | Crystal Structure of SARS CoV-2 Mpro mutant L141R with Pfizer Intravenous Inhibitor PF-00835231 | | 分子名称: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ... | | 著者 | Shaqra, A.M, Schiffer, C.A. | | 登録日 | 2022-07-25 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
8W3M
 
 | | Crystal Structure of Enterovirus 68 3C Protease with AG7404 at 1.97 Angstroms | | 分子名称: | Peptidase C3, SODIUM ION, ethyl (4R)-4-({(2S)-2-[3-{[(5-methyl-1,2-oxazol-3-yl)carbonyl]amino}-2-oxopyridin-1(2H)-yl]pent-4-ynoyl}amino)-5-[(3S)-2-oxopyrrolidin-3-yl]pentanoate | | 著者 | Azzolino, V.N, Shaqra, A.M, Schiffer, C.A. | | 登録日 | 2024-02-22 | | 公開日 | 2025-01-15 | | 最終更新日 | 2025-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Analysis of Inhibitor Binding to Enterovirus-D68 3C Protease.
Viruses, 17, 2025
|
|
8W3C
 
 | |
8W3T
 
 | |
8E7C
 
 | |
3D3T
 
 | | Crystal Structure of HIV-1 CRF01_AE in complex with the substrate p1-p6 | | 分子名称: | HIV-1 protease, P1-P6 SUBSTRATE PEPTIDE | | 著者 | Bandaranayake, R.M, Prabu-Jeyabalan, M, Kakizawa, J, Sugiura, W, Schiffer, C.A. | | 登録日 | 2008-05-12 | | 公開日 | 2008-07-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural analysis of human immunodeficiency virus type 1 CRF01_AE protease in complex with the substrate p1-p6.
J.Virol., 82, 2008
|
|
6WF0
 
 | | Crystal Structure of Broadly Neutralizing Antibody 3I14 Bound to the Influenza A H3 Hemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | 登録日 | 2020-04-03 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.46 Å) | | 主引用文献 | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
6WF1
 
 | | Crystal Structure of Broadly Neutralizing Antibody 3I14 Bound to the Influenza A H10 Hemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | 著者 | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | 登録日 | 2020-04-03 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (4.19 Å) | | 主引用文献 | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
