7LRE
 
 | |
7LRC
 
 | | Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement | | 分子名称: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | 登録日 | 2021-02-16 | | 公開日 | 2021-06-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
6Y20
 
 | |
8R80
 
 | | SARS-CoV-2 Delta RBD in complex with XBB-9 Fab and an anti-Fab nanobody | | 分子名称: | Spike protein S1, XBB-9 Fab heavy chain, XBB-9 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-11-27 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (4.03 Å) | | 主引用文献 | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8R8K
 
 | |
8QRF
 
 | | SARS-CoV-2 delta RBD complexed with XBB-6 and beta-49 Fabs | | 分子名称: | Beta-49 heavy chain, Beta-49 light chain, Spike protein S1, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-10-06 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QTD
 
 | | Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab | | 分子名称: | Spike glycoprotein,Fibritin, XBB-7 fab heavy chain, XBB-7 fab light chain | | 著者 | Ren, J, Duyvesteyn, H.M.E, Stuart, D.I. | | 登録日 | 2023-10-12 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRG
 
 | | SARS-CoV-2 delta RBD complexed with XBB-2 Fab and NbC1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NbC1, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-10-07 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QSQ
 
 | |
3FL7
 
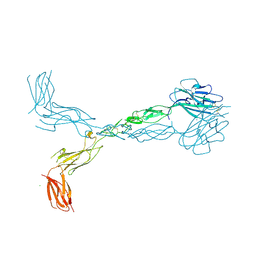 | | Crystal structure of the human ephrin A2 ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Ephrin receptor, ... | | 著者 | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2008-12-18 | | 公開日 | 2009-01-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6UMW
 
 | |
3IIF
 
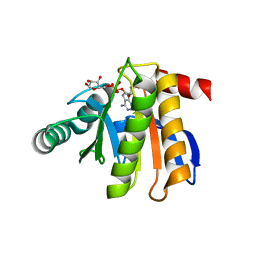 | | Crystal structure of the macro domain of human histone macroH2A1.1 in complex with ADP-ribose (form B) | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Core histone macro-H2A.1, Isoform 1 | | 著者 | Hothorn, M, Bortfeld, M, Ladurner, A.G, Scheffzek, K. | | 登録日 | 2009-07-31 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3IID
 
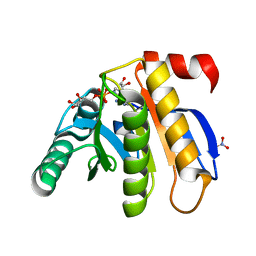 | | Crystal structure of the macro domain of human histone macroH2A1.1 in complex with ADP-ribose (form A) | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Core histone macro-H2A.1, Isoform 1, ... | | 著者 | Hothorn, M, Bortfeld, M, Ladurner, A.G, Scheffzek, K. | | 登録日 | 2009-07-31 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5DL1
 
 | | ClpP from Staphylococcus aureus in complex with AV145 | | 分子名称: | 1-(propan-2-yl)-N-{[2-(thiophen-2-yl)-1,3-oxazol-4-yl]methyl}-1H-pyrazolo[3,4-b]pyridine-5-carboxamide, ATP-dependent Clp protease proteolytic subunit | | 著者 | Vielberg, M.-T, Groll, M. | | 登録日 | 2015-09-04 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Reversible Inhibitors Arrest ClpP in a Defined Conformational State that Can Be Revoked by ClpX Association.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6V02
 
 | | N-terminal 5 domains of CI-MPR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor | | 著者 | Olson, L.J, Dahms, N.M, Kim, J.-J.P. | | 登録日 | 2019-11-18 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Allosteric regulation of lysosomal enzyme recognition by the cation-independent mannose 6-phosphate receptor.
Commun Biol, 3, 2020
|
|
7L28
 
 | | Crystal structure of the catalytic domain of human PDE3A bound to Trequinsin | | 分子名称: | (2E)-9,10-dimethoxy-3-methyl-2-[(2,4,6-trimethylphenyl)imino]-2,3,6,7-tetrahydro-4H-pyrimido[6,1-a]isoquinolin-4-one, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Horner, S.W, Garvie, C. | | 登録日 | 2020-12-16 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L29
 
 | |
7L27
 
 | |
7KWE
 
 | |
4LQM
 
 | | EGFR L858R in complex with PD168393 | | 分子名称: | CHLORIDE ION, Epidermal growth factor receptor, N-[4-(3-BROMO-PHENYLAMINO)-QUINAZOLIN-6-YL]-ACRYLAMIDE | | 著者 | Yun, C.H, Eck, M.J. | | 登録日 | 2013-07-19 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural, Biochemical, and Clinical Characterization of Epidermal Growth Factor Receptor (EGFR) Exon 20 Insertion Mutations in Lung Cancer.
Sci Transl Med, 5, 2013
|
|
4LRM
 
 | | EGFR D770_N771insNPG in complex with PD168393 | | 分子名称: | Epidermal growth factor receptor, N-{4-[(3-bromophenyl)amino]quinazolin-6-yl}propanamide | | 著者 | Yun, C.H, Eck, M.J. | | 登録日 | 2013-07-20 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.526 Å) | | 主引用文献 | Structural, Biochemical, and Clinical Characterization of Epidermal Growth Factor Receptor (EGFR) Exon 20 Insertion Mutations in Lung Cancer.
Sci Transl Med, 5, 2013
|
|
7KPL
 
 | | Crystal structure of hEphB1 in apo form | | 分子名称: | Ephrin type-B receptor 1 | | 著者 | Ahmed, M, Wang, P, Sadek, H. | | 登録日 | 2020-11-11 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.705 Å) | | 主引用文献 | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KPM
 
 | | Crystal structure of hEphB1 bound with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Ephrin type-B receptor 1 | | 著者 | Ahmed, M, Wang, P, Sadek, H. | | 登録日 | 2020-11-11 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.608 Å) | | 主引用文献 | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1SHW
 
 | | EphB2 / EphrinA5 Complex Structure | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 2, Ephrin-A5, ... | | 著者 | Himanen, J.P, Chumley, M.J, Lackmann, M, Li, C, Barton, W.A, Jeffrey, P.D, Vearing, C, Geleick, D, Feldheim, D.A, Boyd, A.W. | | 登録日 | 2004-02-26 | | 公開日 | 2004-05-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling
Nat.Neurosci., 7, 2004
|
|
1SHX
 
 | | Ephrin A5 ligand structure | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin-A5 | | 著者 | Himanen, J.P, Barton, W.A, Nikolov, D.B, Jeffrey, P.D. | | 登録日 | 2004-02-26 | | 公開日 | 2005-04-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Three distinct molecular surfaces in ephrin-A5 are essential for a functional interaction with EphA3.
J.Biol.Chem., 280, 2005
|
|
