6V9O
 
 | |
6FK5
 
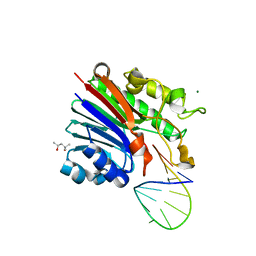 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to DNA substrate in presence of magnesium ion | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), MAGNESIUM ION, ... | | 著者 | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | 登録日 | 2018-01-23 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6VSJ
 
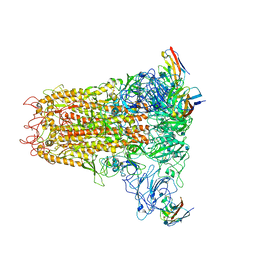 | | Cryo-electron microscopy structure of mouse coronavirus spike protein complexed with its murine receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 1, Spike glycoprotein | | 著者 | Shang, J, Wan, Y.S, Liu, C, Yount, B, Gully, K, Yang, Y, Auerbach, A, Peng, G.Q, Baric, R, Li, F. | | 登録日 | 2020-02-11 | | 公開日 | 2020-03-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.94 Å) | | 主引用文献 | Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry.
Plos Pathog., 16, 2020
|
|
6FKE
 
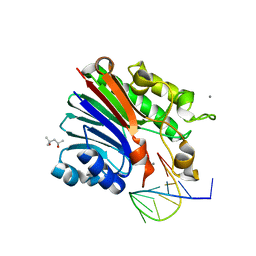 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to product DNA hairpin | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*A)-3'), Exodeoxyribonuclease III, ... | | 著者 | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | 登録日 | 2018-01-23 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.151 Å) | | 主引用文献 | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6FK4
 
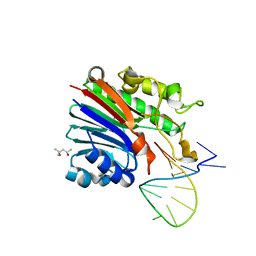 | | Structure of 3' phosphatase NExo (WT) from Neisseria bound to DNA substrate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*GP*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), Exodeoxyribonuclease III | | 著者 | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | 登録日 | 2018-01-23 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6VW1
 
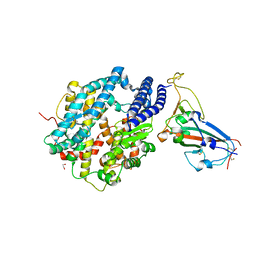 | | Structure of SARS-CoV-2 chimeric receptor-binding domain complexed with its receptor human ACE2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shang, J, Ye, G, Shi, K, Wan, Y.S, Aihara, H, Li, F. | | 登録日 | 2020-02-18 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural basis of receptor recognition by SARS-CoV-2.
Nature, 581, 2020
|
|
1K73
 
 | | Co-crystal Structure of Anisomycin Bound to the 50S Ribosomal Subunit | | 分子名称: | 23S RRNA, 5S RRNA, ANISOMYCIN, ... | | 著者 | Hansen, J, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | 登録日 | 2001-10-18 | | 公開日 | 2003-07-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
1XG7
 
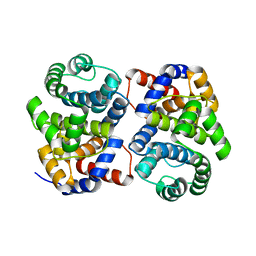 | | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus | | 分子名称: | hypothetical protein | | 著者 | Chang, J, Zhao, M, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Chen, L, Zhou, W, Habel, J, Tempel, W, Lee, D, Lin, D, Chang, S.-H, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Chen, C.-Y, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-09-16 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus
To be published
|
|
5Y6P
 
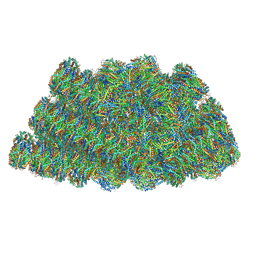 | | Structure of the phycobilisome from the red alga Griffithsia pacifica | | 分子名称: | ApcD, ApcF, LC, ... | | 著者 | Zhang, J, Ma, J.F, Liu, D.S, Sun, S, Sui, S.F. | | 登録日 | 2017-08-13 | | 公開日 | 2017-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of phycobilisome from the red alga Griffithsia pacifica
Nature, 551, 2017
|
|
8K1I
 
 | |
8TBY
 
 | | Apo Bcs1, unsymmetrized | | 分子名称: | Mitochondrial chaperone BCS1 | | 著者 | Zhan, J, Xia, D. | | 登録日 | 2023-06-29 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8T5U
 
 | | ATP-1 state of Bcs1 (C7 symmetrized) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | 著者 | Zhan, J, Xia, D. | | 登録日 | 2023-06-14 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8OIX
 
 | | CryoEM structure of 20S Trichomonas vaginalis proteasome in complex with proteasome inhibitor Salinosporamid A | | 分子名称: | (3AR,6R,6AS)-6-((S)-((S)-CYCLOHEX-2-ENYL)(HYDROXY)METHYL)-6A-METHYL-4-OXO-HEXAHYDRO-2H-FURO[3,2-C]PYRROLE-6-CARBALDEHYDE, Family T1, proteasome alpha subunit, ... | | 著者 | Silhan, J, Fajtova, P, Boura, E. | | 登録日 | 2023-03-23 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Tv20S proteasome in the complex with marizomib
To Be Published
|
|
8T14
 
 | | ADP-bound Bcs1 (C7 symmetrized) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Mitochondrial chaperone BCS1 | | 著者 | Zhan, J, Xia, D. | | 登録日 | 2023-06-01 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8T7U
 
 | | ADP-bound Bcs1 (unsymmetrized) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Mitochondrial chaperone BCS1 | | 著者 | Zhan, J, Xia, D. | | 登録日 | 2023-06-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
3JBU
 
 | | Mechanisms of Ribosome Stalling by SecM at Multiple Elongation Steps | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, J, Pan, X.J, Yan, K.G, Sun, S, Gao, N, Sui, S.F. | | 登録日 | 2015-10-16 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Mechanisms of ribosome stalling by SecM at multiple elongation steps
Elife, 4, 2015
|
|
3JBV
 
 | | Mechanisms of Ribosome Stalling by SecM at Multiple Elongation Steps | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Zhang, J, Pan, X.J, Yan, K.G, Sun, S, Gao, N, Sui, S.F. | | 登録日 | 2015-10-16 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Mechanisms of ribosome stalling by SecM at multiple elongation steps
Elife, 4, 2015
|
|
5SVD
 
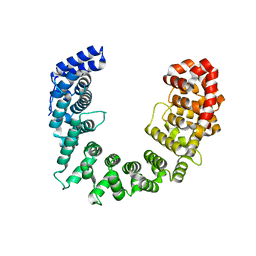 | |
1A4F
 
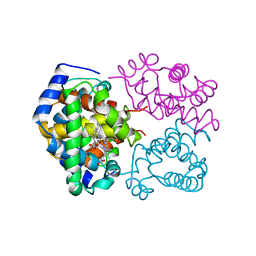 | | BAR-HEADED GOOSE HEMOGLOBIN (OXY FORM) | | 分子名称: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), OXYGEN MOLECULE, ... | | 著者 | Zhang, J, Gu, X. | | 登録日 | 1998-01-29 | | 公開日 | 1998-04-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of a high oxygen affinity species of haemoglobin (bar-headed goose haemoglobin in the oxy form).
J.Mol.Biol., 255, 1996
|
|
5T9P
 
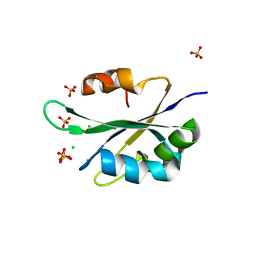 | |
6MCZ
 
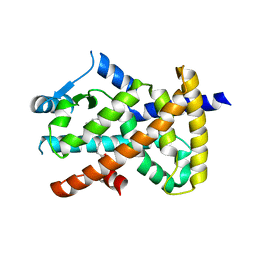 | |
8P0T
 
 | |
6MD4
 
 | |
6MD1
 
 | |
6MD2
 
 | |
