7WD9
 
 | |
7WD7
 
 | |
7WDF
 
 | |
7WD8
 
 | |
7WCR
 
 | |
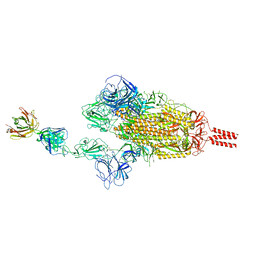
7WCZ
 
 | |
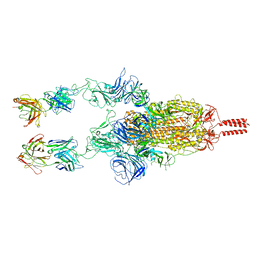
7WD0
 
 | |
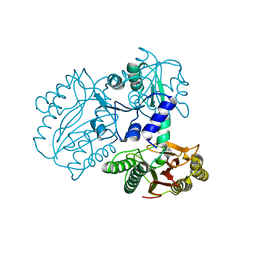
7K3U
 
 | |
6E1A
 
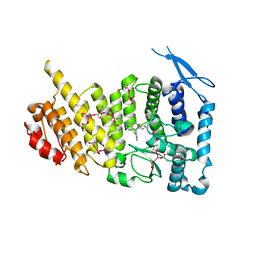 | | Menin bound to M-89 | | 分子名称: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | 著者 | Stuckey, J.A. | | 登録日 | 2018-07-09 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
3C43
 
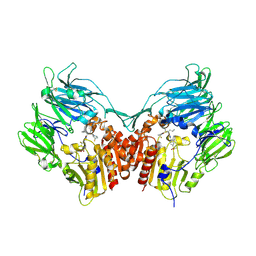 | | Human dipeptidyl peptidase IV/CD26 in complex with a flouroolefin inhibitor | | 分子名称: | (2S,3S)-4-cyclopropyl-3-{(3R,5R)-3-[2-fluoro-4-(methylsulfonyl)phenyl]-1,2,4-oxadiazolidin-5-yl}-1-[(3S)-3-fluoropyrrolidin-1-yl]-1-oxobutan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G, Edmondson, S.D, Weber, A.E. | | 登録日 | 2008-01-29 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Fluoroolefins as amide bond mimics in dipeptidyl peptidase IV inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BM3
 
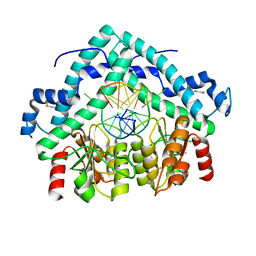 | | Restriction endonuclease PspGI-substrate DNA complex | | 分子名称: | CITRIC ACID, DNA (5'-D(*CP*AP*TP*CP*CP*AP*GP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*CP*CP*TP*GP*GP*AP*T)-3'), ... | | 著者 | Szczepanowski, R.H, Carpenter, M, Czapinska, H, Tamulaitis, G, Siksnys, V, Bhagwat, A, Bochtler, M. | | 登録日 | 2007-12-12 | | 公開日 | 2008-09-16 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A direct crystallographic demonstration that Type II restriction endonuclease PspGI flips nucleotides
To be Published
|
|
4Y2W
 
 | |
7SDC
 
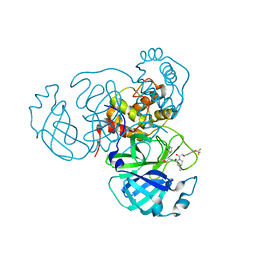 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MI-09 | | 分子名称: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-{[4-(trifluoromethoxy)phenoxy]acetyl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Yang, K.S, Liu, W.R. | | 登録日 | 2021-09-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
7SDA
 
 | |
7SD9
 
 | |
7SH7
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI87 | | 分子名称: | 3C-like proteinase nsp5, benzyl [(2S,3R)-3-tert-butoxy-1-{[(2S)-3-cyclohexyl-1-oxo-1-(2-{[(3S)-2-oxopyrrolidin-3-yl]methyl}-2-propanoylhydrazinyl)propan-2-yl]amino}-1-oxobutan-2-yl]carbamate (non-preferred name) | | 著者 | Blankenship, L.R, Yang, K.S, Liu, W.R. | | 登録日 | 2021-10-08 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | An Azapeptide Platform in Conjunction with Covalent Warheads to Uncover High-Potency Inhibitors for SARS-CoV-2 Main Protease.
Biorxiv, 2023
|
|
5FB7
 
 | |
7TE0
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor PF-07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Yang, K.S, Liu, W.R. | | 登録日 | 2022-01-03 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Evolutionary and Structural Insights about Potential SARS-CoV-2 Evasion of Nirmatrelvir.
J.Med.Chem., 65, 2022
|
|
7YV9
 
 | |
5T03
 
 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and glucuronic acid containing hexasaccharide substrate | | 分子名称: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | 著者 | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | 登録日 | 2016-08-15 | | 公開日 | 2017-02-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5T0A
 
 | | Crystal Structure of Heparan Sulfate 6-O-Sulfotransferase with bound PAP and heptasaccharide substrate | | 分子名称: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | 著者 | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | 登録日 | 2016-08-15 | | 公開日 | 2017-02-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5T05
 
 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and IdoA2S containing hexasaccharide substrate | | 分子名称: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | 著者 | Pedersen, L.C, Moon, A.F, krahn, J.M, Liu, J. | | 登録日 | 2016-08-15 | | 公開日 | 2017-02-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
4N7S
 
 | | Crystal structure of Tse3-Tsi3 complex with Zinc ion | | 分子名称: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Shang, G.J. | | 登録日 | 2013-10-16 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4N88
 
 | | Crystal structure of Tse3-Tsi3 complex with calcium ion | | 分子名称: | CALCIUM ION, Uncharacterized protein | | 著者 | Shang, G.J. | | 登録日 | 2013-10-17 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
4N80
 
 | | Crystal structure of Tse3-Tsi3 complex | | 分子名称: | CALCIUM ION, Uncharacterized protein, ZINC ION | | 著者 | Shang, G.J. | | 登録日 | 2013-10-16 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
