2AIX
 
 | |
7U1Z
 
 | | Crystal structure of the DRBD and CROPs of TcdA | | 分子名称: | SULFATE ION, Toxin A | | 著者 | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | 登録日 | 2022-02-22 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
7U2P
 
 | | Structure of TcdA GTD in complex with RhoA | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Glucosyltransferase TcdA, MAGNESIUM ION, ... | | 著者 | Baohua, C, Zheng, L, Kay, P, Rongsheng, J. | | 登録日 | 2022-02-24 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.596 Å) | | 主引用文献 | Structure of the glucosyltransferase domain of TcdA in complex with RhoA provides insights into substrate recognition.
Sci Rep, 12, 2022
|
|
7UBX
 
 | |
1XHY
 
 | | X-ray structure of the Y702F mutant of the GluR2 ligand-binding core (S1S2J) in complex with kainate at 1.85 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor, SULFATE ION | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-09-21 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
7UBY
 
 | | Structure of the GTD domain of Clostridium difficile toxin A in complex with VHH AH3 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Glucosyltransferase TcdA, ... | | 著者 | Chen, B, Rongsheng, J, Kay, P. | | 登録日 | 2022-03-15 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Neutralizing epitopes on Clostridioides difficile toxin A revealed by the structures of two camelid VHH antibodies.
Front Immunol, 13, 2022
|
|
6O9U
 
 | | KirBac3.1 at a resolution of 2 Angstroms | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3,3',3''-phosphoryltripropanoic acid, BARIUM ION, ... | | 著者 | Gulbis, J.M, Clarke, O.B. | | 登録日 | 2019-03-15 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
6O9T
 
 | | KirBac3.1 mutant at a resolution of 4.1 Angstroms | | 分子名称: | 2,3,5,6-tetramethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Inward rectifier potassium channel Kirbac3.1, POTASSIUM ION | | 著者 | Gulbis, J.M, Black, K.A, Miller, D.M. | | 登録日 | 2019-03-15 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.01 Å) | | 主引用文献 | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
6O9V
 
 | | KirBac3.1 mutant at a resolution of 3.1 Angstroms | | 分子名称: | 1,1-Methanediyl Bismethanethiosulfonate, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Inward rectifier potassium channel Kirbac3.1, ... | | 著者 | Gulbis, J.M, Black, K.A, Miller, D.M. | | 登録日 | 2019-03-15 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.094 Å) | | 主引用文献 | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
7S0Z
 
 | | Structures of TcdB in complex with R-Ras | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | 著者 | Zheng, L, Rongsheng, J, Peng, C. | | 登録日 | 2021-08-31 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structural basis for selective modification of Rho and Ras GTPases by Clostridioides difficile toxin B.
Sci Adv, 7, 2021
|
|
7S0Y
 
 | | Structures of TcdB in complex with Cdc42 | | 分子名称: | Cell division control protein 42 homolog, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Zheng, L, Rongsheng, J, Peng, C. | | 登録日 | 2021-08-31 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural basis for selective modification of Rho and Ras GTPases by Clostridioides difficile toxin B.
Sci Adv, 7, 2021
|
|
3H6H
 
 | |
3H6G
 
 | |
8WRH
 
 | | SARS-CoV-2 XBB.1.5.70 in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | 著者 | Feng, L.L, Feng, L.L. | | 登録日 | 2023-10-14 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRL
 
 | | XBB.1.5 RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | 著者 | Feng, L.L, Feng, L.L. | | 登録日 | 2023-10-15 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRO
 
 | | XBB.1.5.10 spike protein in complex with ACE2 | | 分子名称: | Processed angiotensin-converting enzyme 2, Spike glycoprotein,Spike glycoprotein,Spike glycoprotein,Fusion protein | | 著者 | Feng, L.L, Feng, L.L. | | 登録日 | 2023-10-15 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRM
 
 | | XBB.1.5 spike protein in complex with ACE2 | | 分子名称: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Feng, L.L, Feng, L.L. | | 登録日 | 2023-10-15 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.34 Å) | | 主引用文献 | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTJ
 
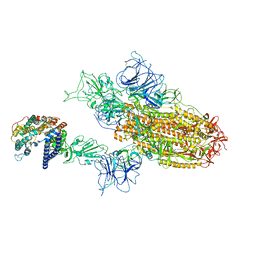 | | XBB.1.5.70 spike protein in complex with ACE2 | | 分子名称: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Feng, L.L, Feng, L.L. | | 登録日 | 2023-10-18 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.64 Å) | | 主引用文献 | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WTD
 
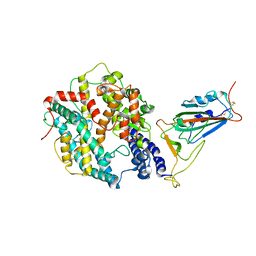 | | XBB.1.5.10 RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | 著者 | Feng, L.L, Feng, L.L. | | 登録日 | 2023-10-18 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
1FTJ
 
 | |
8XRY
 
 | |
8XNG
 
 | |
8XW3
 
 | |
8XS4
 
 | |
8XW1
 
 | |
