3ZIW
 
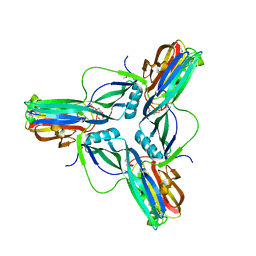 | | Clostridium perfringens enterotoxin, D48A mutation and N-terminal 37 residues deleted | | 分子名称: | HEAT-LABILE ENTEROTOXIN B CHAIN, HEXAETHYLENE GLYCOL | | 著者 | Yelland, T, Naylor, C.E, Savva, C.G, Basak, A.K. | | 登録日 | 2013-01-14 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a C. Perfringens Enterotoxin Mutant in Complex with a Modified Claudin-2 Extracellular Loop 2
J.Mol.Biol., 426, 2014
|
|
1XJU
 
 | |
1XJT
 
 | |
1QFL
 
 | |
7K9S
 
 | | Cryptococcus neoformans Hsp90 nucleotide binding domain in complex with NVP-AUY922 | | 分子名称: | 5-[2,4-DIHYDROXY-5-(1-METHYLETHYL)PHENYL]-N-ETHYL-4-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]ISOXAZOLE-3-CARBOXAMIDE, Hsp90-like protein | | 著者 | Kuntz, D.A, Prive, G.G. | | 登録日 | 2020-09-29 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Fungal-Selective Resorcylate Aminopyrazole Hsp90 Inhibitors: Optimization of Whole-Cell Anticryptococcal Activity and Insights into the Structural Origins of Cryptococcal Selectivity.
J.Med.Chem., 64, 2021
|
|
1KTA
 
 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE : THREE DIMENSIONAL STRUCTURE OF THE ENZYME IN ITS PYRIDOXAMINE PHOSPHATE FORM. | | 分子名称: | 3-METHYL-2-OXOBUTANOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ACETIC ACID, ... | | 著者 | Yennawar, N.H, Conway, M.E, Yennawar, H.P, Farber, G.K, Hutson, S.M. | | 登録日 | 2002-01-15 | | 公開日 | 2002-11-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of human mitochondrial branched chain aminotransferase reaction intermediates: ketimine and pyridoxamine phosphate forms
Biochemistry, 41, 2002
|
|
7K9R
 
 | |
7K9W
 
 | | Cryptococcus neoformans Hsp90 nucleotide binding domain in complex with BUCMD00461 | | 分子名称: | (1,3-dihydro-2H-isoindol-2-yl)(2,4-dihydroxy-6-{[3-(3-methoxyphenyl)-1-methyl-1H-pyrazol-5-yl]amino}phenyl)methanone, BENZAMIDINE, CHLORIDE ION, ... | | 著者 | Kuntz, D.A, Prive, G.G. | | 登録日 | 2020-09-29 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Fungal-Selective Resorcylate Aminopyrazole Hsp90 Inhibitors: Optimization of Whole-Cell Anticryptococcal Activity and Insights into the Structural Origins of Cryptococcal Selectivity.
J.Med.Chem., 64, 2021
|
|
1RKB
 
 | | The structure of adrenal gland protein AD-004 | | 分子名称: | LITHIUM ION, Protein AD-004, SULFATE ION | | 著者 | Ren, H, Liang, Y, Bennett, M, Su, X.D. | | 登録日 | 2003-11-21 | | 公開日 | 2005-01-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of human adenylate kinase 6: An adenylate kinase localized to the cell nucleus
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7K9U
 
 | | Cryptococcus neoformans Hsp90 nucleotide binding domain in complex with BUCMD00429 | | 分子名称: | (5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl){2,4-dihydroxy-6-[(1-methyl-3-phenyl-1H-pyrazol-5-yl)amino]phenyl}methanone, Hsp90-like protein | | 著者 | Kuntz, D.A, Prive, G.G. | | 登録日 | 2020-09-29 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Fungal-Selective Resorcylate Aminopyrazole Hsp90 Inhibitors: Optimization of Whole-Cell Anticryptococcal Activity and Insights into the Structural Origins of Cryptococcal Selectivity.
J.Med.Chem., 64, 2021
|
|
7K9V
 
 | | Cryptococcus neoformans Hsp90 nucleotide binding domain in complex with BUCMD00452 | | 分子名称: | (1,3-dihydro-2H-isoindol-2-yl)(2,4-dihydroxy-6-{[1-methyl-3-(2-methylphenyl)-1H-pyrazol-5-yl]amino}phenyl)methanone, BENZAMIDINE, CHLORIDE ION, ... | | 著者 | Kuntz, D.A, Prive, G.G. | | 登録日 | 2020-09-29 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Fungal-Selective Resorcylate Aminopyrazole Hsp90 Inhibitors: Optimization of Whole-Cell Anticryptococcal Activity and Insights into the Structural Origins of Cryptococcal Selectivity.
J.Med.Chem., 64, 2021
|
|
1QNI
 
 | | Crystal Structure of Nitrous Oxide Reductase from Pseudomonas nautica, at 2.4A Resolution | | 分子名称: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Brown, K, Tegoni, M, Cambillau, C. | | 登録日 | 1999-10-15 | | 公開日 | 2000-10-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A novel type of catalytic copper cluster in nitrous oxide reductase.
Nat.Struct.Biol., 7, 2000
|
|
1EKV
 
 | | HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL): THREE DIMENSIONAL STRUCTURE OF ENZYME INACTIVATED BY TRIS BOUND TO THE PYRIDOXAL-5'-PHOSPHATE ON ONE END AND ACTIVE SITE LYS202 NZ ON THE OTHER. | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | 著者 | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | 登録日 | 2000-03-09 | | 公開日 | 2001-03-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EKP
 
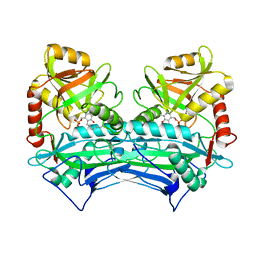 | | CRYSTAL STRUCTURE OF HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL) COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE AT 2.5 ANGSTROMS (MONOCLINIC FORM). | | 分子名称: | BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | 著者 | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | 登録日 | 2000-03-09 | | 公開日 | 2001-03-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1EKF
 
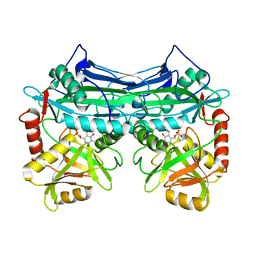 | | CRYSTALLOGRAPHIC STRUCTURE OF HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL) COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE AT 1.95 ANGSTROMS (ORTHORHOMBIC FORM) | | 分子名称: | BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | 著者 | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | 登録日 | 2000-03-08 | | 公開日 | 2001-03-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HB7
 
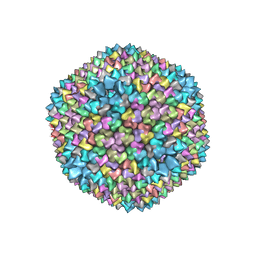 | | quasi-atomic resolution model of bacteriophage PRD1 sus1 mutant, obtained by combined cryo-EM and X-ray crystallography. | | 分子名称: | BACTERIOPHAGE PRD1 SUS1 MUTANT CAPSID | | 著者 | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | 登録日 | 2001-04-12 | | 公開日 | 2001-12-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (14 Å) | | 主引用文献 | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
1HB9
 
 | | quasi-atomic resolution model of bacteriophage PRD1 wild type virion, obtained by combined cryo-EM and X-ray crystallography. | | 分子名称: | BACTERIOPHAGE PRD1 | | 著者 | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | 登録日 | 2001-04-13 | | 公開日 | 2001-12-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (25 Å) | | 主引用文献 | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions
Structure, 9, 2001
|
|
1GUB
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | 分子名称: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, NICKEL (II) ION | | 著者 | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | 登録日 | 2002-01-24 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | 分子名称: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | 著者 | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | 登録日 | 2002-01-24 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
2M4X
 
 | |
2M4Z
 
 | |
1CTF
 
 | |
2L2I
 
 | |
2M50
 
 | | Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin, Huwentoxin-IV (-TRTX-Hh2a). | | 分子名称: | Mu-theraphotoxin-Hh2a | | 著者 | Gibbs, A, Minassian, N, Flinspach, M, Wickenden, A. | | 登録日 | 2013-02-12 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Analysis of the Structural and Molecular Basis of Voltage-sensitive Sodium Channel Inhibition by the Spider Toxin Huwentoxin-IV ( mu-TRTX-Hh2a).
J.Biol.Chem., 288, 2013
|
|
2OFS
 
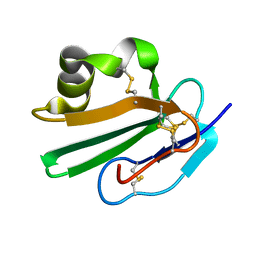 | | Crystal structure of human CD59 | | 分子名称: | CD59 glycoprotein | | 著者 | Davies, C. | | 登録日 | 2007-01-04 | | 公開日 | 2007-05-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structure of CD59: implications for molecular recognition of the complement proteins C8 and C9 in the membrane-attack complex.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
