2KB6
 
 | | Solution structure of onconase C87A/C104A | | 分子名称: | Protein P-30 | | 著者 | Weininger, U, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Balbach, J. | | 登録日 | 2008-11-21 | | 公開日 | 2009-11-24 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
7NXA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 B.1.351 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXB
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 P.1 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXC
 
 | |
7NX6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab Heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX9
 
 | | Crystal structure of the N501Y mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX7
 
 | | Crystal structure of the K417N mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX8
 
 | | Crystal structure of the K417T mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
3JZF
 
 | |
3QDA
 
 | |
1GFF
 
 | |
3SUC
 
 | |
2YI7
 
 | | Structural characterization of 5-Aryl-4-(5-substituted-2-4- dihydroxyphenyl)-1,2,3-thiadiazole Hsp90 inhibitors. | | 分子名称: | 4-CHLORO-6-[5-(4-ETHOXYPHENYL)-1,2,3-THIADIAZOL-4-YL BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION | | 著者 | Roe, S.M, Prodromou, C, Pearl, L.H. | | 登録日 | 2011-05-10 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Co-Crystalization and in Vitro Biological Characterization of 5-Aryl-4-(5-Substituted-2-4-Dihydroxyphenyl)-1,2,3-Thiadiazole Hsp90 Inhibitors.
Plos One, 7, 2012
|
|
3KYR
 
 | | Bace-1 in complex with a norstatine type inhibitor | | 分子名称: | 3-[[(2S)-2-[[[(2S)-2-[[(2S)-2-[[(2S)-2-azanyl-3-(1H-1,2,3,4-tetrazol-5-ylcarbonylamino)propanoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-pentanoyl]amino]methyl]-2-hydroxy-4-phenyl-butanoyl]amino]benzoic acid, Beta-secretase 1 | | 著者 | Lindberg, J.D, Borkakoti, N, Derbyshire, D, Nystrom, S. | | 登録日 | 2009-12-07 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Investigation of a-phenylnorstatine and a-benzylnorstatine as transition state isostere motifs in the search for new BACE-1 inhibiotrs
To be Published
|
|
2F98
 
 | | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. | | 分子名称: | Aklanonic Acid methyl Ester Cyclase, AknH, METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, ... | | 著者 | Kallio, P, Sultana, A, Neimi, J, Mantsala, P, Schneider, G. | | 登録日 | 2005-12-05 | | 公開日 | 2006-02-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity.
J.Mol.Biol., 357, 2006
|
|
7S0E
 
 | |
7S0D
 
 | |
7S0B
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | 著者 | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2021-08-30 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
7S0C
 
 | |
3OMK
 
 | |
2F99
 
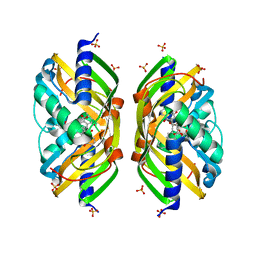 | | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity. | | 分子名称: | Aklanonic Acid methyl Ester Cyclase, AknH, SULFATE ION, ... | | 著者 | Kallio, P, Sultana, A, Neimi, J, Mantsala, P, Schneider, G. | | 登録日 | 2005-12-05 | | 公開日 | 2006-02-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the polyketide cyclase AknH with bound substrate and product analogue: implications for catalytic mechanism and product stereoselectivity.
J.Mol.Biol., 357, 2006
|
|
7A6Y
 
 | |
4OK9
 
 | |
3HBR
 
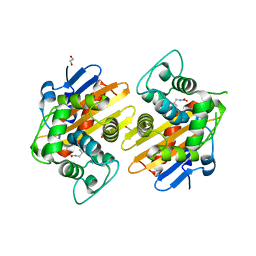 | | Crystal structure of OXA-48 beta-lactamase | | 分子名称: | 1,2-ETHANEDIOL, OXA-48 | | 著者 | Calderone, V, Mangani, S, Benvenuti, M, Rossolini, G.M, Docquier, J.D. | | 登録日 | 2009-05-05 | | 公開日 | 2009-06-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the OXA-48 beta-lactamase reveals mechanistic diversity among class D carbapenemases.
Chem.Biol., 16, 2009
|
|
7A6R
 
 | |
