4IRR
 
 | | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thymidylate synthase | | 著者 | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | 登録日 | 2013-01-15 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP
To be Published
|
|
4ISW
 
 | | Crystal Structure of Phosphorylated C.elegans Thymidylate Synthase in Complex with dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | 著者 | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | 登録日 | 2013-01-17 | | 公開日 | 2013-12-11 | | 最終更新日 | 2014-01-15 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Crystal structure of phosphoramide-phosphorylated thymidylate synthase reveals pSer127, reflecting probably pHis to pSer phosphotransfer.
Bioorg.Chem., 52C, 2013
|
|
3AYU
 
 | | Crystal structure of MMP-2 active site mutant in complex with APP-drived decapeptide inhibitor | | 分子名称: | 72 kDa type IV collagenase, Amyloid beta A4 protein, CALCIUM ION, ... | | 著者 | Hashimoto, H, Takeuchi, T, Komatsu, K, Miyazaki, K, Sato, M, Higashi, S. | | 登録日 | 2011-05-17 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for matrix metalloproteinase-2 (MMP-2)-selective inhibitory action of {beta}-amyloid precursor protein-derived inhibitor
J.Biol.Chem., 2011
|
|
4JWV
 
 | | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444 | | 分子名称: | Short chain enoyl-CoA hydratase | | 著者 | Cooper, D.R, Mikolajczak, K, Cymborowski, M, Grabowski, M, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-03-27 | | 公開日 | 2013-05-29 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of putative short chain enoyl-CoA hydratase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
4JYL
 
 | | Crystal structure of enoyl-CoA hydratase from Thermoplasma volcanium GSS1 | | 分子名称: | CHLORIDE ION, Enoyl-CoA hydratase, SULFATE ION | | 著者 | Shabalin, I.G, Cooper, D.R, Majorek, K.A, Mikolajczak, K, Porebski, P.J, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-03-29 | | 公開日 | 2013-04-17 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Crystal structure of enoyl-CoA hydratase from Thermoplasma volcanium GSS1
To be Published
|
|
2DGA
 
 | | Crystal structure of hexameric beta-glucosidase in wheat | | 分子名称: | Beta-glucosidase, GLYCEROL, SULFATE ION | | 著者 | Sue, M, Yamazaki, K, Miyamoto, T, Yajima, S. | | 登録日 | 2006-03-10 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular and Structural Characterization of Hexameric beta-D-Glucosidases in Wheat and Rye.
Plant Physiol., 141, 2006
|
|
1UHT
 
 | | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein | | 分子名称: | expressed protein | | 著者 | Tomizawa, T, Inoue, M, Koshiba, S, Hayashi, F, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Shinozaki, K, Seki, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-07-10 | | 公開日 | 2004-01-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of The FHA Domain of Arabidopsis thaliana Hypothetical Protein
To be Published
|
|
1G0C
 
 | | ALKALINE CELLULASE K CATALYTIC DOMAIN-CELLOBIOSE COMPLEX | | 分子名称: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE, ... | | 著者 | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | 登録日 | 2000-10-05 | | 公開日 | 2001-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
1G01
 
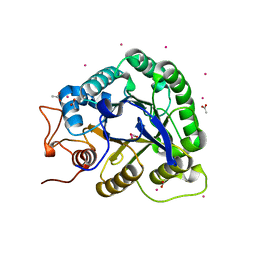 | | ALKALINE CELLULASE K CATALYTIC DOMAIN | | 分子名称: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE | | 著者 | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | 登録日 | 2000-10-05 | | 公開日 | 2001-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
2ZWN
 
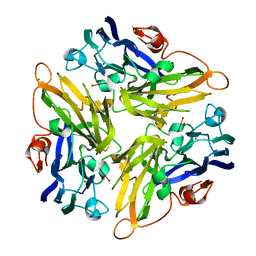 | | Crystal structure of the novel two-domain type laccase from a metagenome | | 分子名称: | CHLORIDE ION, COPPER (II) ION, CU-O-CU LINKAGE, ... | | 著者 | Komori, H, Miyazaki, K, Higuchi, Y. | | 登録日 | 2008-12-17 | | 公開日 | 2009-04-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray structure of a two-domain type laccase: a missing link in the evolution of multi-copper proteins
Febs Lett., 583, 2009
|
|
6R2N
 
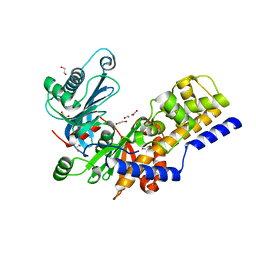 | | Crystal structure of KlGlk1 glucokinase from Kluyveromyces lactis | | 分子名称: | 1,2-ETHANEDIOL, BROMIDE ION, Glucokinase-1 | | 著者 | Zak, K, Wator, E, Grudnik, P. | | 登録日 | 2019-03-18 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.596 Å) | | 主引用文献 | Crystal Structure of Kluyveromyces lactis Glucokinase ( Kl Glk1).
Int J Mol Sci, 20, 2019
|
|
8TG8
 
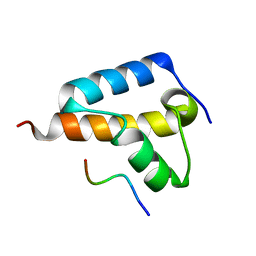 | |
8TFU
 
 | |
8TGC
 
 | |
8TG7
 
 | |
7Z0J
 
 | | human PEX13 SH3 domain in complex with internal FxxxF motif | | 分子名称: | 1,2-ETHANEDIOL, Peroxisomal membrane protein PEX13 | | 著者 | Gaussmann, S, Zak, K, Kreisz, N, Sattler, M. | | 登録日 | 2022-02-23 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Intramolecular autoinhibition of human PEX13 modulates peroxisomal import
Biorxiv, 2022
|
|
7Z0I
 
 | | human PEX13 SH3 domain | | 分子名称: | 1,2-ETHANEDIOL, Peroxisomal membrane protein PEX13, ZINC ION | | 著者 | Gaussmann, S, Zak, K, Sattler, M. | | 登録日 | 2022-02-23 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Intramolecular autoinhibition of human PEX13 modulates peroxisomal import
Biorxiv, 2022
|
|
6R73
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed meropenem | | 分子名称: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, ZINC ION | | 著者 | Softley, C.A, Zak, K, Kolonko, M, Sattler, M, Popowicz, G. | | 登録日 | 2019-03-28 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
8R41
 
 | | Structure of CHI3L1 in complex with inhibitor 1 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | 著者 | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | 登録日 | 2023-11-10 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
8R4X
 
 | | Structure of Chitinase-3-like protein 1 in complex with inhibitor 30 | | 分子名称: | (2~{S},5~{S})-4-[1-(4-chloranylpyridin-2-yl)piperidin-4-yl]-5-[(4-chlorophenyl)methyl]-2-methyl-morpholine, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | 登録日 | 2023-11-14 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
8R42
 
 | | Structure of CHI3L1 in complex with inhibititor 2 | | 分子名称: | 1,2-ETHANEDIOL, 2-[4-[(2~{R})-2-[(4-chlorophenyl)methyl]pyrrolidin-1-yl]piperidin-1-yl]pyridine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | 登録日 | 2023-11-10 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
8AUP
 
 | | Structure of hARG1 with a novel inhibitor. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[(1~{R},3~{R},4~{S})-3-azanyl-3-carboxy-4-[(dimethylamino)methyl]cyclohexyl]ethyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, ... | | 著者 | Napiorkowska-Gromadzka, A, Nowak, E, Nowotny, M. | | 登録日 | 2022-08-25 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Arginase 1/2 Inhibitor OATD-02: From Discovery to First-in-man Setup in Cancer Immunotherapy.
Mol.Cancer Ther., 22, 2023
|
|
1IDM
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE, LOOP-DELETED CHIMERA | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Sakurai, M, Ohzeki, M, Moriyama, H, Sato, M, Tanaka, N. | | 登録日 | 1995-05-19 | | 公開日 | 1995-09-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a loop-deleted variant of 3-isopropylmalate dehydrogenase from Thermus thermophilus: an internal reprieve tolerance mechanism.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7BV9
 
 | | The NMR structure of the BEN domain from human NAC1 | | 分子名称: | Nucleus accumbens-associated protein 1 | | 著者 | Nagata, T, Kobayashi, N, Nakayama, N, Obayashi, E, Urano, T. | | 登録日 | 2020-04-09 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nucleus Accumbens-Associated Protein 1 Binds DNA Directly through the BEN Domain in a Sequence-Specific Manner.
Biomedicines, 8, 2020
|
|
3JRS
 
 | | Crystal structure of (+)-ABA-bound PYL1 | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At5g46790 | | 著者 | Miyazono, K, Miyakawa, T, Sawano, Y, Kubota, K, Tanokura, M. | | 登録日 | 2009-09-08 | | 公開日 | 2009-11-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis of abscisic acid signalling
Nature, 462, 2009
|
|
