6QK9
 
 | |
4GEH
 
 | |
6AVO
 
 | | Cryo-EM structure of human immunoproteasome with a novel noncompetitive inhibitor that selectively inhibits activated lymphocytes | | 分子名称: | N~1~-{2-[([1,1'-biphenyl]-3-carbonyl)amino]ethyl}-N~4~-tert-butyl-N~2~-(3-phenylpropanoyl)-L-aspartamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | 著者 | Li, H, Santos, R, Bai, L. | | 登録日 | 2017-09-04 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of human immunoproteasome with a reversible and noncompetitive inhibitor that selectively inhibits activated lymphocytes.
Nat Commun, 8, 2017
|
|
6OB1
 
 | |
4YVQ
 
 | |
5B76
 
 | |
7YGG
 
 | |
8JSO
 
 | | AMPH-bound hTAAR1-Gs protein complex | | 分子名称: | (2S)-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | 登録日 | 2023-06-20 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6B70
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin | | 分子名称: | FAB H11-E heavy chain, FAB H11-E light chain, Insulin, ... | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-03 | | 公開日 | 2017-12-27 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
8JSP
 
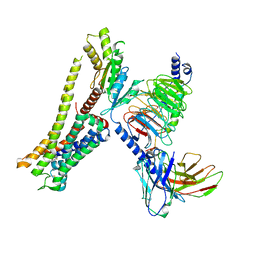 | | Ulotaront(SEP-363856)-bound Serotonin 1A (5-HT1A) receptor-Gi complex | | 分子名称: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, 5-hydroxytryptamine receptor 1A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | 登録日 | 2023-06-20 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
4XT9
 
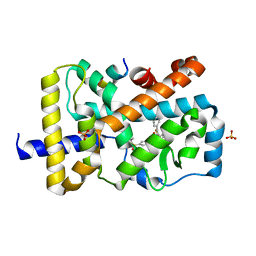 | | RORgamma (263-509) complexed with GSK2435341A and SRC2 | | 分子名称: | LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, N-[4-(2,5-dichlorophenyl)-5-phenyl-1,3-thiazol-2-yl]-2-[4-(ethylsulfonyl)phenyl]acetamide, Nuclear receptor ROR-gamma, ... | | 著者 | Wang, Y, Ma, Y. | | 登録日 | 2015-01-23 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Discovery of N-(4-aryl-5-aryloxy-thiazol-2-yl)-amides as potent ROR gamma t inverse agonists
Bioorg.Med.Chem., 23, 2015
|
|
6VZB
 
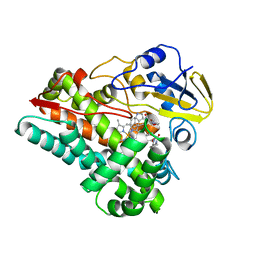 | | Crystal structure of cytochrome P450 NasF5053 S284A-V288A mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | 分子名称: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | 著者 | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | 登録日 | 2020-02-28 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6BF8
 
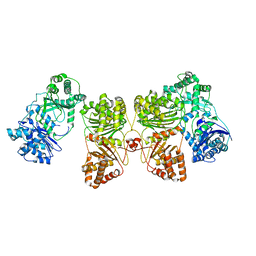 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Y
 
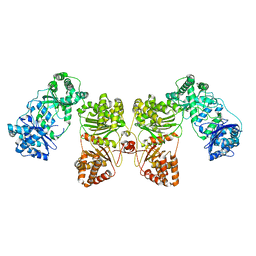 | | Cryo-EM structure of human insulin degrading enzyme | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6VXV
 
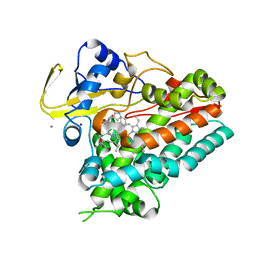 | | Crystal structure of cyclo-L-Trp-L-Pro-bound cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | 分子名称: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | 登録日 | 2020-02-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6W0S
 
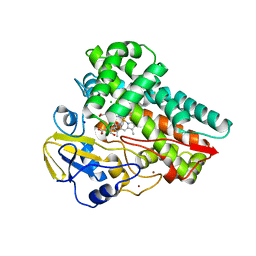 | | Crystal structure of substrate free cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | 分子名称: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | 登録日 | 2020-03-02 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6BFC
 
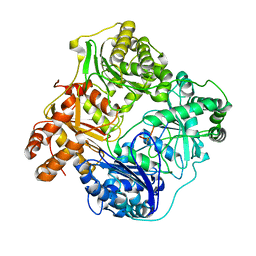 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2017-12-27 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
5CHE
 
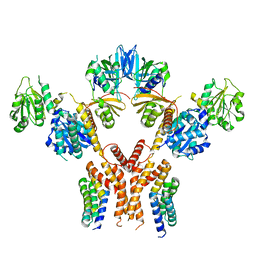 | |
6BF6
 
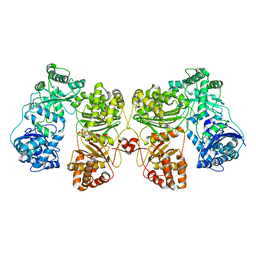 | | Cryo-EM structure of human insulin degrading enzyme | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B3Q
 
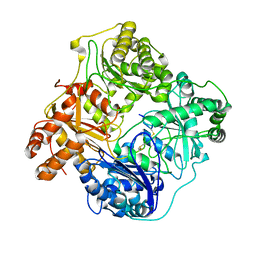 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-09-22 | | 公開日 | 2017-11-22 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6B7Z
 
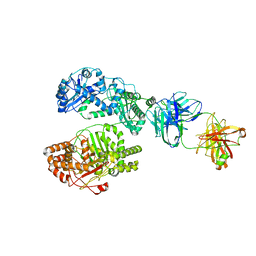 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11 heavy chain and FAB H11 light chain | | 分子名称: | FAB H11 heavy chain, FAB H11 light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2018-01-10 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
4ZHW
 
 | |
4ZHY
 
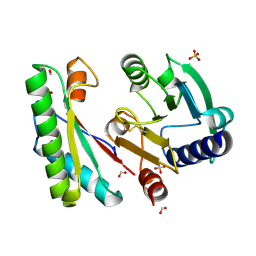 | | Crystal structure of a bacterial signalling complex | | 分子名称: | FORMIC ACID, SULFATE ION, YfiB, ... | | 著者 | Li, S, Li, T, Wang, Y, Bartlam, M. | | 登録日 | 2015-04-27 | | 公開日 | 2016-04-27 | | 実験手法 | X-RAY DIFFRACTION (1.969 Å) | | 主引用文献 | Structural insights into YfiR sequestering by YfiB in Pseudomonas aeruginosa PAO1
Sci Rep, 5, 2015
|
|
