4XV1
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX7904 | | 分子名称: | N'-(3-{[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)-N-ethyl-N-methylsulfuric diamide, Serine/threonine-protein kinase B-raf | | 著者 | Zhang, Y, Zhang, C. | | 登録日 | 2015-01-26 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | RAF inhibitors that evade paradoxical MAPK pathway activation.
Nature, 526, 2015
|
|
8END
 
 | |
8ENF
 
 | |
8ENB
 
 | |
3LBL
 
 | | Structure of human MDM2 protein in complex with Mi-63-analog | | 分子名称: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | 登録日 | 2010-01-08 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
8EEQ
 
 | | CryoEM structures of bAE1 captured in multiple states. | | 分子名称: | Anion exchange protein | | 著者 | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | 登録日 | 2022-09-07 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
3LBK
 
 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | 分子名称: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | 登録日 | 2010-01-08 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
4FZF
 
 | | Crystal structure of MST4-MO25 complex with DKI | | 分子名称: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZD
 
 | | Crystal structure of MST4-MO25 complex with WSF motif | | 分子名称: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
3LBJ
 
 | | Structure of human MDMX protein in complex with a small molecule inhibitor | | 分子名称: | N-[(3S)-1-({6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indol-2-yl}carbonyl)pyrrolidin-3-yl]-N,N',N'-trimethylpropane-1,3-diamine, Protein Mdm4, SULFATE ION | | 著者 | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | 登録日 | 2010-01-08 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7YRZ
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Zhou, Y.R, Zeng, P, Zhou, X.L, Lin, C, Zhang, J, Yin, X.S, Li, J. | | 登録日 | 2022-08-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
4FZA
 
 | | Crystal structure of MST4-MO25 complex | | 分子名称: | Calcium-binding protein 39, GLYCEROL, Serine/threonine-protein kinase MST4 | | 著者 | Shi, Z.B, Zhou, Z.C. | | 登録日 | 2012-07-06 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4RP9
 
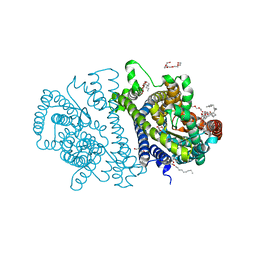 | | Bacterial vitamin C transporter UlaA/SgaT in C2 form | | 分子名称: | ASCORBIC ACID, Ascorbate-specific permease IIC component UlaA, PENTAETHYLENE GLYCOL, ... | | 著者 | Wang, J.W. | | 登録日 | 2014-10-29 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Crystal structure of a phosphorylation-coupled vitamin C transporter.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RP8
 
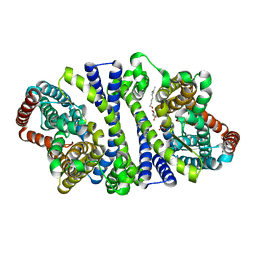 | | Bacterial vitamin C transporter UlaA/SgaT in P21 form | | 分子名称: | ASCORBIC ACID, Ascorbate-specific permease IIC component UlaA, nonyl beta-D-glucopyranoside | | 著者 | Wang, J.W. | | 登録日 | 2014-10-29 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.359 Å) | | 主引用文献 | Crystal structure of a phosphorylation-coupled vitamin C transporter.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4JH0
 
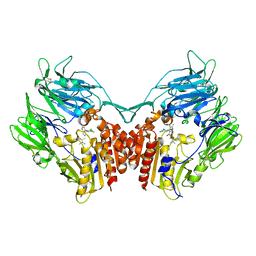 | | Crystal structure of dipeptidyl-peptidase 4 (CD26, adenosine deaminase complexing protein 2) (DPP-IV-WT) complex with bms-767778 AKA 2-(3-(aminomethyl)-4-(2,4- dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6h-pyrrolo[3,4- b]pyridin-6-yl)-n,n-dimethylacetamide | | 分子名称: | 2-[3-(aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5,7-dihydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-N,N-dimethylacetamide, Dipeptidyl peptidase 4 | | 著者 | Klei, H.E. | | 登録日 | 2013-03-04 | | 公開日 | 2013-09-04 | | 最終更新日 | 2013-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Optimization of Activity, Selectivity, and Liability Profiles in 5-Oxopyrrolopyridine DPP4 Inhibitors Leading to Clinical Candidate (Sa)-2-(3-(Aminomethyl)-4-(2,4-dichlorophenyl)-2-methyl-5-oxo-5H-pyrrolo[3,4-b]pyridin-6(7H)-yl)-N,N-dimethylacetamide (BMS-767778).
J.Med.Chem., 56, 2013
|
|
8WKJ
 
 | |
8WOU
 
 | |
5CFF
 
 | |
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | 分子名称: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | 著者 | Xu, Z.H, Zhou, Z. | | 登録日 | 2020-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | 分子名称: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | 著者 | Xu, Z.H, Zhou, Z. | | 登録日 | 2020-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.254 Å) | | 主引用文献 | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
5BNQ
 
 | | Crystal structure of hRANKL-mRANK complex | | 分子名称: | CHLORIDE ION, PHOSPHATE ION, SODIUM ION, ... | | 著者 | Ren, J. | | 登録日 | 2015-05-26 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A RANKL mutant used as an inter-species vaccine for efficient immunotherapy of osteoporosis.
Sci Rep, 5, 2015
|
|
7E23
 
 | | SARS-CoV-2 spike in complex with the CA521 neutralizing antibody Fab (focused refinement on Fab-RBD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CA521 Heavy Chain, CA521 Light Chain, ... | | 著者 | Liu, C, Song, D, Dou, C. | | 登録日 | 2021-02-04 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure and function analysis of a potent human neutralizing antibody CA521 FALA against SARS-CoV-2.
Commun Biol, 4, 2021
|
|
8XJ3
 
 | |
