7DHJ
 
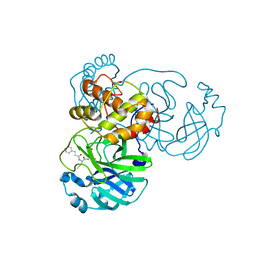 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | 分子名称: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | 著者 | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | 登録日 | 2020-11-15 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.962 Å) | | 主引用文献 | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
7DGF
 
 | |
7DGG
 
 | |
7DGI
 
 | |
7VOP
 
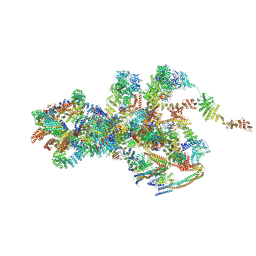 | | Cryo-EM structure of Xenopus laevis nuclear pore complex cytoplasmic ring subunit | | 分子名称: | GATOR complex protein SEC13, IL4I1 protein, MGC154553 protein, ... | | 著者 | Tai, L, Zhu, Y, Sun, F. | | 登録日 | 2021-10-14 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (8.7 Å) | | 主引用文献 | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7VCI
 
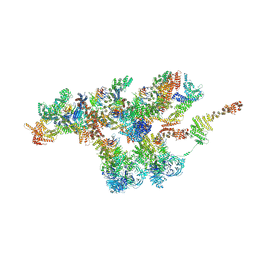 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | 分子名称: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | 著者 | Tai, L, Zhu, Y, Sun, F. | | 登録日 | 2021-09-03 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (8.1 Å) | | 主引用文献 | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7Y9U
 
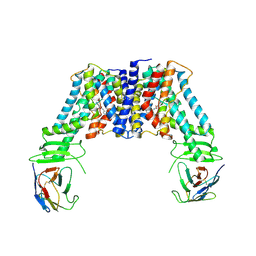 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the NPA-bound state | | 分子名称: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 1, nanobody | | 著者 | Sun, L, Liu, X, Yang, Z, Xia, J. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-07 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y9T
 
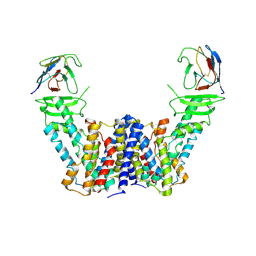 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the apo state | | 分子名称: | Auxin efflux carrier component 1, nanobody | | 著者 | Sun, L, Liu, X, Yang, Z, Xia, J. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-07 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
3HO2
 
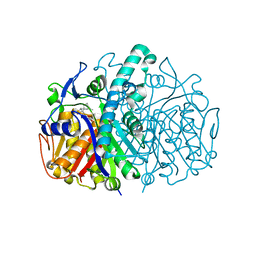 | | Structure of E.coli FabF(C163A) in complex with Platencin | | 分子名称: | 2,4-dihydroxy-3-({3-[(2S,4aS,8S,8aR)-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonaphthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | 著者 | Soisson, S.M, Parthasarathy, G. | | 登録日 | 2009-06-01 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Isolation, enzyme-bound structure and antibacterial activity of platencin A1 from Streptomyces platensis.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6M3M
 
 | |
6JCA
 
 | |
6JC8
 
 | |
3I8P
 
 | |
3HO9
 
 | | Structure of E.coli FabF(C163A) in complex with Platencin A1 | | 分子名称: | 2,4-dihydroxy-3-({3-[(2R,4aR,8S,8aR,9R)-9-hydroxy-8-methyl-3-methylidene-7-oxo-1,3,4,7,8,8a-hexahydro-2H-2,4a-ethanonap hthalen-8-yl]propanoyl}amino)benzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2 | | 著者 | Soisson, S.M, Parthasarathy, G. | | 登録日 | 2009-06-01 | | 公開日 | 2010-02-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Isolation, enzyme-bound structure and antibacterial activity of platencin A1 from Streptomyces platensis.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HNZ
 
 | |
2L6G
 
 | |
7WK5
 
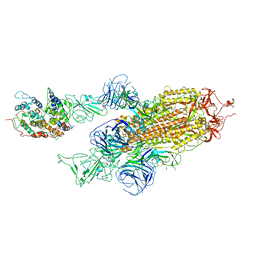 | | Cryo-EM structure of Omicron S-ACE2, C2 state | | 分子名称: | Angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Han, W.Y, Wang, Y.F. | | 登録日 | 2022-01-08 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-03-09 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization
Biorxiv, 2022
|
|
7WK3
 
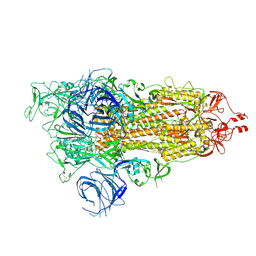 | | SARS-CoV-2 Omicron S-open | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-01-08 | | 公開日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
6BL9
 
 | |
3NPZ
 
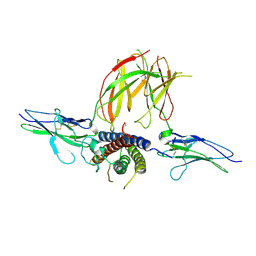 | | Prolactin Receptor (PRLR) Complexed with the Natural Hormone (PRL) | | 分子名称: | Prolactin, Prolactin receptor | | 著者 | Van Agthoven, J, England, P, Goffin, V, Broutin, I. | | 登録日 | 2010-06-29 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structural characterization of the stem-stem dimerization interface between prolactin receptor chains complexed with the natural hormone.
J.Mol.Biol., 404, 2010
|
|
4GJT
 
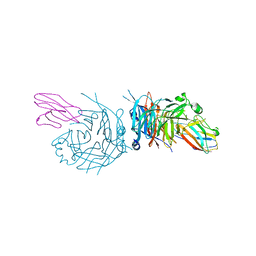 | | complex structure of nectin-4 bound to MV-H | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Poliovirus receptor-related protein 4 | | 著者 | Zhang, X, Lu, G, Qi, J, Li, Y, He, Y, Xu, X, Shi, J, Zhang, C, Yan, J, Gao, G.F. | | 登録日 | 2012-08-10 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1001 Å) | | 主引用文献 | Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nat.Struct.Mol.Biol., 20, 2013
|
|
7XKE
 
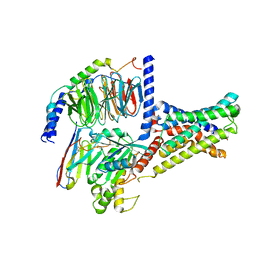 | | Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex | | 分子名称: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | 登録日 | 2022-04-19 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKF
 
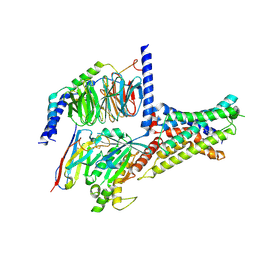 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state | | 分子名称: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | 登録日 | 2022-04-19 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKD
 
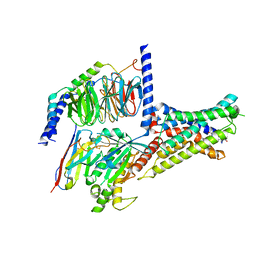 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex | | 分子名称: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | 登録日 | 2022-04-19 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
3N94
 
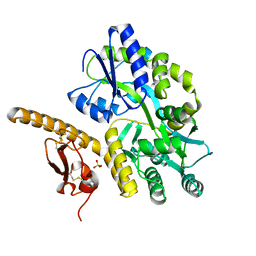 | | Crystal structure of human pituitary adenylate cyclase 1 Receptor-short N-terminal extracellular domain | | 分子名称: | Fusion protein of Maltose-binding periplasmic protein and pituitary adenylate cyclase 1 Receptor-short, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Kumar, S, Pioszak, A.A, Swaminathan, K, Xu, H.E. | | 登録日 | 2010-05-28 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the PAC1R Extracellular Domain Unifies a Consensus Fold for Hormone Recognition by Class B G-Protein Coupled Receptors.
Plos One, 6, 2011
|
|
