7Q72
 
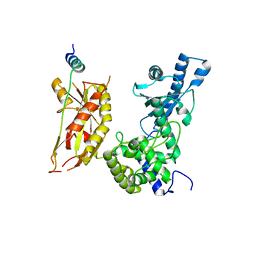 | | Structure of Pla1 in complex with Red1 | | 分子名称: | NURS complex subunit red1, Poly(A) polymerase pla1 | | 著者 | Soni, K, Wild, K, Sinning, I. | | 登録日 | 2021-11-09 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q73
 
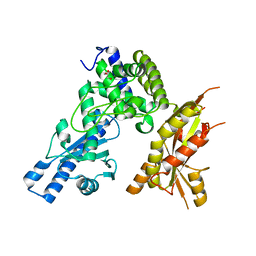 | | Structure of Pla1 apo | | 分子名称: | GLYCEROL, Poly(A) polymerase pla1 | | 著者 | Soni, K, Wild, K, Sinning, I. | | 登録日 | 2021-11-09 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q74
 
 | |
7EA6
 
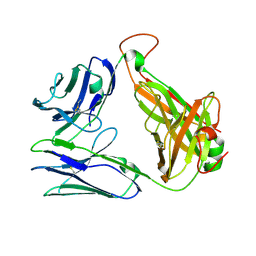 | | Crystal structure of TCR-017 ectodomain | | 分子名称: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | 著者 | Nagae, M, Yamasaki, S. | | 登録日 | 2021-03-06 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.18000245 Å) | | 主引用文献 | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
6UNQ
 
 | | Kinase domain of ALK2-K493A with AMPPNP | | 分子名称: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Agnew, C, Jura, N. | | 登録日 | 2019-10-13 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNR
 
 | | Kinase domain of ALK2-K492A/K493A with AMPPNP | | 分子名称: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Agnew, C, Jura, N. | | 登録日 | 2019-10-13 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNP
 
 | | Crystal structure of the kinase domain of BMPR2-D485G | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bone morphogenetic protein receptor type-2, ... | | 著者 | Agnew, C, Jura, N. | | 登録日 | 2019-10-13 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNS
 
 | | Kinase domain of ALK2-K492A/K493A with LDN-193189 | | 分子名称: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1 | | 著者 | Agnew, C, Jura, N. | | 登録日 | 2019-10-13 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
7YOW
 
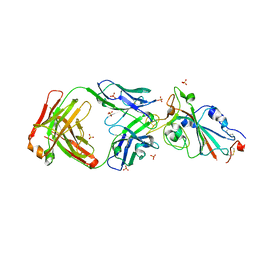 | |
7DEQ
 
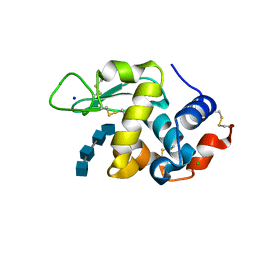 | | Lysozyme-sugar complex in D2O | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Tanaka, I, Chatake, T. | | 登録日 | 2020-11-04 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DER
 
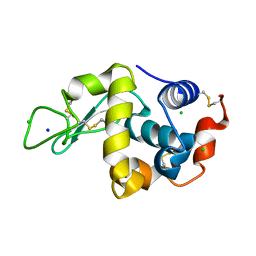 | | Lysozyme alone in H2O | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Tanaka, I, Chatake, T. | | 登録日 | 2020-11-04 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7BR5
 
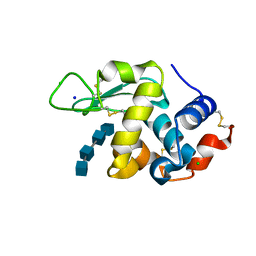 | | Lysozyme-sugar complex in H2O | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Tanaka, I, Chatake, T. | | 登録日 | 2020-03-26 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4Q9K
 
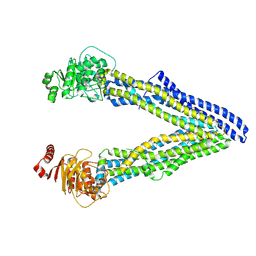 | | P-glycoprotein cocrystallised with QZ-Leu | | 分子名称: | (30F)L(30F)L(30F)L Peptide, Multidrug resistance protein 1A | | 著者 | McGrath, A.P, Szewczyk, P, Chang, G. | | 登録日 | 2014-05-01 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9I
 
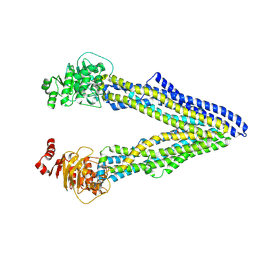 | | P-glycoprotein cocrystallised with QZ-Ala | | 分子名称: | (30F)A(30F)A(30F)A Peptide, Multidrug resistance protein 1A | | 著者 | McGrath, A.P, Szewczyk, P, Chang, G. | | 登録日 | 2014-05-01 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (3.781 Å) | | 主引用文献 | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9J
 
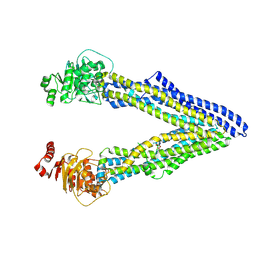 | | P-glycoprotein cocrystallised with QZ-Val | | 分子名称: | Multidrug resistance protein 1A, N-{[5-AMINO-1-(5-O-PHOSPHONO-BETA-D-ARABINOFURANOSYL)-1H-IMIDAZOL-4-YL]CARBONYL}-L-ASPARTIC ACID | | 著者 | McGrath, A.P, Szewczyk, P, Chang, G. | | 登録日 | 2014-05-01 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9H
 
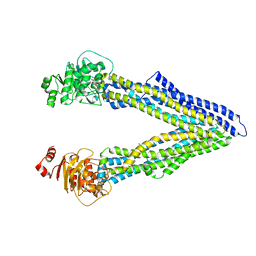 | |
4DIR
 
 | |
4Q9L
 
 | | P-glycoprotein cocrystallised with QZ-Phe | | 分子名称: | (30F)F(30F)F(30F)F Peptide, Multidrug resistance protein 1A | | 著者 | McGrath, A.P, Szewczyk, P, Chang, G. | | 登録日 | 2014-05-01 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3BRK
 
 | |
8H78
 
 | | Crystal structure of human MMP-2 catalytic domain in complex with inhibitor | | 分子名称: | (2~{R})-2-[[4-[(4-aminocarbonylphenyl)carbonylamino]phenyl]sulfonylamino]-5-[(2~{S},4~{S})-4-azanyl-2-[[(2~{S})-1-[[(2~{S})-1-[(5-azanyl-5-oxidanylidene-pentyl)amino]-5-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]-methyl-amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamoyl]pyrrolidin-1-yl]-5-oxidanylidene-pentanoic acid, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | 著者 | Kamitani, M, Takeuchi, T, Mima, M. | | 登録日 | 2022-10-19 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Discovery of TP0597850: A Selective, Chemically Stable, and Slow Tight-Binding Matrix Metalloproteinase-2 Inhibitor with a Phenylbenzamide-Pentapeptide Hybrid Scaffold.
J.Med.Chem., 66, 2023
|
|
6L0V
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NGR2, ... | | 著者 | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | 登録日 | 2019-09-27 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.347 Å) | | 主引用文献 | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
6L0W
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | 分子名称: | 1,2-ETHANEDIOL, CITRATE ANION, NGR2, ... | | 著者 | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | 登録日 | 2019-09-27 | | 公開日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.591 Å) | | 主引用文献 | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
6KB0
 
 | | X-ray structure of human PPARalpha ligand binding domain-5,8,11,14-eicosatetraynoic acid (ETYA) co-crystals obtained by soaking | | 分子名称: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, icosa-5,8,11,14-tetraynoic acid | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB1
 
 | | X-ray structure of human PPARalpha ligand binding domain-tetradecylthioacetic acid (TTA) co-crystals obtained by soaking | | 分子名称: | 2-tetradecylsulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KAZ
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by soaking | | 分子名称: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | 著者 | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | 登録日 | 2019-06-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
