7MEJ
 
 | |
7MDW
 
 | |
7ME7
 
 | |
6LZZ
 
 | | Crystal structure of the PDE9 catalytic domain in complex with inhibitor 4a | | 分子名称: | 1-cyclopentyl-6-[[(2R)-1-(2-oxa-6-azaspiro[3.3]heptan-6-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Huang, Y.Y, Wu, Y, Luo, H.B. | | 登録日 | 2020-02-19 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.40003753 Å) | | 主引用文献 | Identification of phosphodiesterase-9 as a novel target for pulmonary arterial hypertension by using highly selective and orally bioavailable inhibitors
To Be Published
|
|
5Y37
 
 | | Crystal structure of GBS GAPDH | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Jin, T, Zhou, K. | | 登録日 | 2017-07-28 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | High-resolution crystal structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
1RKB
 
 | | The structure of adrenal gland protein AD-004 | | 分子名称: | LITHIUM ION, Protein AD-004, SULFATE ION | | 著者 | Ren, H, Liang, Y, Bennett, M, Su, X.D. | | 登録日 | 2003-11-21 | | 公開日 | 2005-01-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of human adenylate kinase 6: An adenylate kinase localized to the cell nucleus
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6N33
 
 | | Crystal structure of fms kinase domain with a small molecular inhibitor, PLX5622 | | 分子名称: | 6-fluoro-N-[(5-fluoro-2-methoxypyridin-3-yl)methyl]-5-[(5-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | 著者 | Zhang, Y. | | 登録日 | 2018-11-14 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer's disease model.
Nat Commun, 10, 2019
|
|
4DRF
 
 | |
4ZBN
 
 | |
5F9P
 
 | | Crystal structure study of anthrone oxidase-like protein | | 分子名称: | Anthrone oxidase-like protein, GLYCEROL | | 著者 | Gao, X, Wu, D, Fan, K, Liu, Z.-J. | | 登録日 | 2015-12-10 | | 公開日 | 2016-12-14 | | 最終更新日 | 2018-07-18 | | 実験手法 | X-RAY DIFFRACTION (2.078 Å) | | 主引用文献 | Structure and Function of a C-C Bond Cleaving Oxygenase in Atypical Angucycline Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
2EXX
 
 | | Crystal structure of HSCARG from Homo sapiens in complex with NADP | | 分子名称: | GLYCEROL, HSCARG protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Dai, X, Chen, Q, Yao, D, Liang, Y, Dong, Y, Gu, X, Zheng, X, Luo, M. | | 登録日 | 2005-11-09 | | 公開日 | 2006-11-21 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Restructuring of the dinucleotide-binding fold in an NADP(H) sensor protein.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2GKD
 
 | |
6JCB
 
 | |
6JLC
 
 | |
7WVO
 
 | | SARS-CoV-2 Omicron S-open-2 | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-02-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
8IUO
 
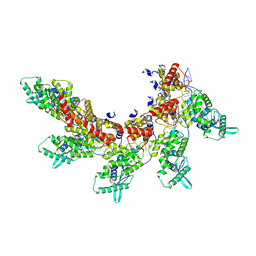 | | respiratory syncytial virus nucleocapsid-like assembly | | 分子名称: | Nucleoprotein, RNA (35-MER) | | 著者 | Wang, Y, Luo, Y, Ling, X, Luo, B, Jia, G, Dong, H, Su, Z. | | 登録日 | 2023-03-24 | | 公開日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Cryo-EM structure of the nucleocapsid-like assembly of respiratory syncytial virus.
Signal Transduct Target Ther, 8, 2023
|
|
7WVN
 
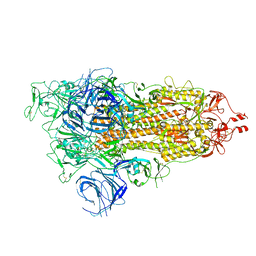 | | SARS-CoV-2 Omicron S-open | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-02-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
2L6H
 
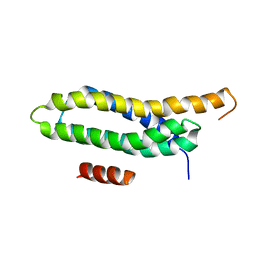 | |
8JPX
 
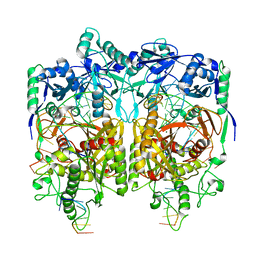 | |
2L6F
 
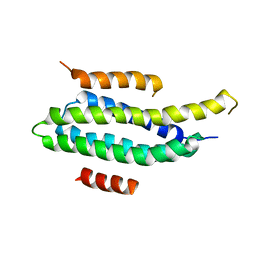 | | NMR Solution structure of FAT domain of FAK complexed with LD2 and LD4 motifs of PAXILLIN | | 分子名称: | Focal adhesion kinase 1, linker1, Paxillin, ... | | 著者 | Bertolucci, C.M, Guibao, C, Zhang, C, Zheng, J. | | 登録日 | 2010-11-19 | | 公開日 | 2012-05-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Solution Structure of Fat Domain of Fak Complexed with Ld2 and Ld4 Motifs of Paxillin
To be Published
|
|
5ZTC
 
 | |
7Y9V
 
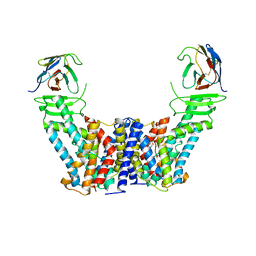 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the IAA-bound state | | 分子名称: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 1, nanobody | | 著者 | Sun, L, Liu, X, Yang, Z, Xia, J. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-07 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y4H
 
 | |
7DGB
 
 | |
7DGH
 
 | |
