4IU6
 
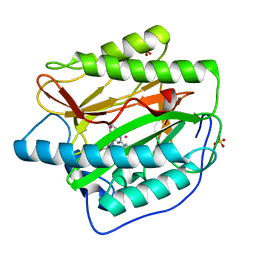 | | Human Methionine Aminopeptidase in complex with FZ1: Pyridinylquinazolines Selectively Inhibit Human Methionine Aminopeptidase-1 | | 分子名称: | 4-[4-(4-methoxyphenyl)piperazin-1-yl]-2-(pyridin-2-yl)quinazoline, COBALT (II) ION, Methionine aminopeptidase 1, ... | | 著者 | Gabelli, S.B, Zhang, F, Miller, M, Liu, J, Amzel, L.M. | | 登録日 | 2013-01-19 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pyridinylquinazolines selectively inhibit human methionine aminopeptidase-1 in cells.
J.Med.Chem., 56, 2013
|
|
5UMO
 
 | |
5XXY
 
 | |
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | 分子名称: | Pachytene checkpoint protein 2 homolog | | 著者 | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | 登録日 | 2019-12-17 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
8T2I
 
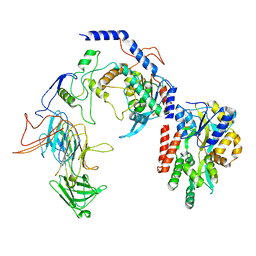 | | Negative stain EM assembly of MYC, JAZ, and NINJA complex | | 分子名称: | AFP homolog 2, Maltose/maltodextrin-binding periplasmic protein, Protein TIFY 10A, ... | | 著者 | Zhou, X.E, Zhang, Y, Zhou, Y, He, Q, Cao, X, Kariapper, L, Suino-Powell, K, Zhu, Y, Zhang, F, Karsten, M. | | 登録日 | 2023-06-06 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (10.4 Å) | | 主引用文献 | Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Plant Commun., 4, 2023
|
|
3CYJ
 
 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | 分子名称: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | 著者 | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-04-25 | | 公開日 | 2008-05-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
3DIP
 
 | | Crystal structure of an enolase protein from the environmental genome shotgun sequencing of the Sargasso Sea | | 分子名称: | SULFATE ION, enolase | | 著者 | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-06-20 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of an enolase protein from the environmental genome shotgun sequencing of the Sargasso Sea
To be Published
|
|
4U63
 
 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | 著者 | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | 登録日 | 2014-07-26 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
6T2P
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16 | | 著者 | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | 登録日 | 2019-10-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
6T2O
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16 | | 著者 | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | 登録日 | 2019-10-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
6T2S
 
 | | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown | | 分子名称: | Glycoside hydrolase family 16 protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | 著者 | Crouch, L.I, Liberato, M.V, Ubranowicz, P.A, Basle, A, Lamb, C.A, Cooke, K, Doona, M, Needham, S, Brady, R.R, Berrington, J.E, Madubic, K, Chater, P, Zhang, F, Linhardt, R.J, Spence, D.I.R, Bolam, D.N. | | 登録日 | 2019-10-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-08-26 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Prominent members of the human gut microbiota express endo-acting O-glycanases to initiate mucin breakdown.
Nat Commun, 11, 2020
|
|
9CEX
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 4 | | 分子名称: | DNA (29-MER), DNA (5'-D(*(MG)*(MG)P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF2
 
 | | Parasitella parasitica Fanzor (PpFz) State 3 | | 分子名称: | DNA non-target strand, DNA substrate model, DNA target strand, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9C0I
 
 | |
8W1P
 
 | |
9CEU
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 1 | | 分子名称: | DNA (5'-D(P*CP*CP*TP*AP*TP*AP*GP*AP*TP*AP*TP*GP*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), Maltose/maltodextrin-binding periplasmic protein,Spizellomyces punctatus Fanzor 1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF1
 
 | | Parasitella parasitica Fanzor (PpFz) State 2 | | 分子名称: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CES
 
 | | Guillardia theta Fanzor (GtFz) State 2 | | 分子名称: | DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*GP*GP*GP*GP*CP*CP*TP*TP*TP*AP*AP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEW
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 3 | | 分子名称: | DNA (29-MER), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF3
 
 | | Parasitella parasitica Fanzor (PpFz) State 4 | | 分子名称: | DNA non-target strand, DNA target strand, MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF0
 
 | | Parasitella parasitica Fanzor (PpFz) State 1 | | 分子名称: | DNA non-target strand, DNA target strand, Maltose/maltodextrin-binding periplasmic protein,Parasitella parasitica Fanzor 1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CER
 
 | | Guillardia theta Fanzor (GtFz) State 1 | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, RNA (142-MER), ZINC ION | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CET
 
 | | Guillardia theta Fanzor (GtFz) State 3 | | 分子名称: | DNA (28-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*TP*AP*AP*AP*GP*GP*CP*CP*CP*CP*GP*GP*G)-3'), Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEV
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 2 | | 分子名称: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CEY
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 5 | | 分子名称: | DNA (26-MER), DNA (36-MER), MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2024-06-27 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
