8JF4
 
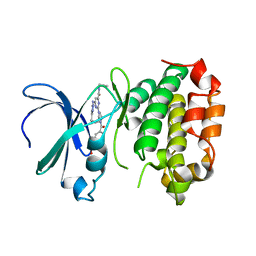 | | The crystal structure of human AURKA kinase domain in complex with AURKA-compound 9 | | 分子名称: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Aurora kinase A | | 著者 | Zhu, C.J. | | 登録日 | 2023-05-17 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.89288354 Å) | | 主引用文献 | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8JF3
 
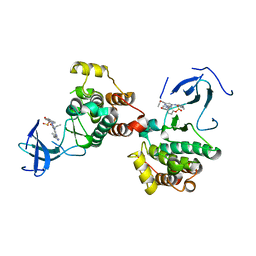 | | C-Src in complex with compound 9 | | 分子名称: | 2-[4-[4-[bis(oxidanylidene)-$l^5-sulfanyl]oxyphenyl]carbonylpiperazin-1-yl]-6-[(5-cyclopropyl-1H-pyrazol-3-yl)amino]-N-prop-2-ynyl-pyrimidine-4-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Zhang, Z.M, Huang, H.S. | | 登録日 | 2023-05-17 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.84647632 Å) | | 主引用文献 | Global Reactivity Profiling of the Catalytic Lysine in Human Kinome for Covalent Inhibitor Development.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8JG8
 
 | |
8JTN
 
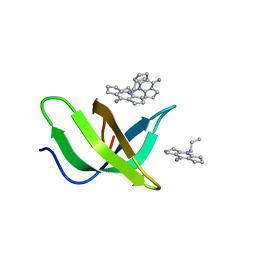 | | Tudor domain of TDRD3 in complex with a small molecule | | 分子名称: | 2-propyl-2-azoniatricyclo[7.3.0.0^{3,7}]dodeca-1(9),2,7-trien-8-amine, Tudor domain-containing protein 3 | | 著者 | Chen, M, Wang, Z, Li, W, Shang, X, Liu, Y. | | 登録日 | 2023-06-22 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Tudor domain of TDRD3 in complex with a small molecule antagonist.
Biochim Biophys Acta Gene Regul Mech, 1866, 2023
|
|
7DL9
 
 | |
7DLA
 
 | |
7DWO
 
 | |
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | 著者 | Wang, W.W, Wu, H, He, J.H, Yu, F. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
7EPW
 
 | |
7EPV
 
 | |
7WN1
 
 | | Structure of PfNT1(Y190A) in complex with nanobody 48 and inosine | | 分子名称: | Equilibrative nucleoside/nucleobase transporter, INOSINE, nanobody48 | | 著者 | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | 登録日 | 2022-01-17 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7YDQ
 
 | | Structure of PfNT1(Y190A)-GFP in complex with GSK4 | | 分子名称: | 5-methyl-N-[2-(2-oxidanylideneazepan-1-yl)ethyl]-2-phenyl-1,3-oxazole-4-carboxamide, Nucleoside transporter 1,Green fluorescent protein | | 著者 | Wang, C, Yu, L.Y, Li, J.L, Ren, R.B, Deng, D. | | 登録日 | 2022-07-04 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7YKC
 
 | |
7WN0
 
 | | Structure of PfENT1(Y190A) in complex with nanobody 19 | | 分子名称: | Equilibrative nucleoside/nucleobase transporter, nanobody19 | | 著者 | Wang, C, Deng, D, Ren, R.B, Yu, L.Y. | | 登録日 | 2022-01-17 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Structural basis of the substrate recognition and inhibition mechanism of Plasmodium falciparum nucleoside transporter PfENT1.
Nat Commun, 14, 2023
|
|
7DIY
 
 | | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-exoribonuclease domain | | 分子名称: | MAGNESIUM ION, ZINC ION, nsp10 protein, ... | | 著者 | Lin, S, Chen, H, Chen, Z.M, Yang, F.L, Ye, F, Zheng, Y, Yang, J, Lin, X, Sun, H.L, Wang, L.L, Wen, A, Cao, Y, Lu, G.W. | | 登録日 | 2020-11-19 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity.
Nucleic Acids Res., 49, 2021
|
|
7E27
 
 | | Structure of PfFNT in complex with MMV007839 | | 分子名称: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | 著者 | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | 登録日 | 2021-02-04 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.29 Å) | | 主引用文献 | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
7E26
 
 | | Structure of PfFNT in apo state | | 分子名称: | Formate-nitrite transporter | | 著者 | Yan, C.Y, Jiang, X, Deng, D, Peng, X, Wang, N, Zhu, A, Xu, H, Li, J. | | 登録日 | 2021-02-04 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.29 Å) | | 主引用文献 | Structural characterization of the Plasmodium falciparum lactate transporter PfFNT alone and in complex with antimalarial compound MMV007839 reveals its inhibition mechanism.
Plos Biol., 19, 2021
|
|
6IGZ
 
 | | Structure of PSI-LHCI | | 分子名称: | (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | 著者 | Xiong, P, Xiaochun, Q. | | 登録日 | 2018-09-27 | | 公開日 | 2019-02-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits.
Nat Plants, 5, 2019
|
|
4ECC
 
 | | Chimeric GST Containing Inserts of Kininogen Peptides | | 分子名称: | chimeric protein between GSHKT10 and domain 5 of kininogen-1 | | 著者 | Amber, A.B, Sergei, M.M, Yi, P, Rita, R, Xiaoping, Q, Marianne, P.-C, William, C.M, Vivien, Y, Keith, R.M, Anton, A.K. | | 登録日 | 2012-03-26 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Chimeric glutathione S-transferases containing inserts of kininogen peptides: potential novel protein therapeutics.
J.Biol.Chem., 287, 2012
|
|
4ECB
 
 | | Chimeric GST Containing Inserts of Kininogen Peptides | | 分子名称: | chimeric protein between GSHKT10 and domain 5 of kininogen-1 | | 著者 | Amber, A.B, Sergei, M.M, Yi, P, Rita, R, Xiaoping, Q, Marianne, P.-C, William, C.M, Vivien, Y, Keith, R.M, Anton, A.K. | | 登録日 | 2012-03-26 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Chimeric glutathione S-transferases containing inserts of kininogen peptides: potential novel protein therapeutics.
J.Biol.Chem., 287, 2012
|
|
