5ZLA
 
 | | Crystal structure of mutant C387A of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | 分子名称: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase C387A mutant | | 著者 | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | 登録日 | 2018-03-27 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKU
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 in complex with DFA-III | | 分子名称: | (2R,3'S,4'S,4aR,5'R,6R,7R,7aS)-4a,5',6-tris(hydroxymethyl)spiro[3,6,7,7a-tetrahydrofuro[2,3-b][1,4]dioxine-2,2'-oxolane ]-3',4',7-triol, DFA-IIIase | | 著者 | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | 登録日 | 2018-03-26 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKS
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 | | 分子名称: | DFA-IIIase | | 著者 | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | 登録日 | 2018-03-26 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZKY
 
 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 without its lid | | 分子名称: | DFA-IIIase | | 著者 | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | 登録日 | 2018-03-26 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
7XIJ
 
 | | Crystal structure of CBP bromodomain liganded with Y08175 | | 分子名称: | 3-[(1-ethanoyl-5-methoxy-indol-3-yl)carbonylamino]-4-fluoranyl-5-(1-methylpyrazol-4-yl)benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | 登録日 | 2022-04-13 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Crystal structure of CBP bromodomain liganded with Y08175
To Be Published
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | 分子名称: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | 著者 | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | 登録日 | 2022-04-28 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
7W9P
 
 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV pi helix conformer) | | 分子名称: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | 登録日 | 2021-12-10 | | 公開日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9M
 
 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (S6IV pi helix conformer) | | 分子名称: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | 登録日 | 2021-12-10 | | 公開日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7WJU
 
 | | Cryo-EM structure of the AsCas12f1-sgRNAv1-dsDNA ternary complex | | 分子名称: | Non-target strand, OrfB_Zn_ribbon domain-containing protein, Target strand, ... | | 著者 | Wu, Z, Liu, D, Shen, H, Ji, Q. | | 登録日 | 2022-01-07 | | 公開日 | 2023-01-18 | | 実験手法 | ELECTRON MICROSCOPY (2.69 Å) | | 主引用文献 | Structure-directed functional evolution of the miniature CRISPR-AsCas12f1 system
To Be Published
|
|
7W9K
 
 | | Cryo-EM structure of human Nav1.7-beta1-beta2 complex at 2.2 angstrom resolution | | 分子名称: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | 登録日 | 2021-12-09 | | 公開日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9L
 
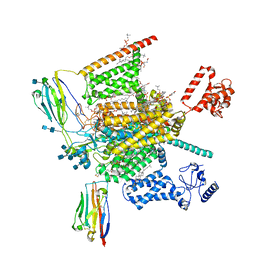 | | Cryo-EM structure of human Nav1.7(E406K)-beta1-beta2 complex | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | 登録日 | 2021-12-10 | | 公開日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | 分子名称: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | 著者 | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | 登録日 | 2021-05-21 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
6A90
 
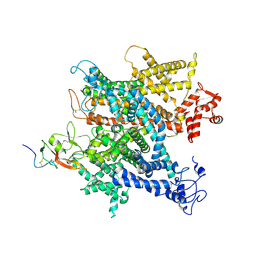 | | Complex of voltage-gated sodium channel NavPaS from American cockroach Periplaneta americana and Dc1a | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mu-diguetoxin-Dc1a, ... | | 著者 | Shen, H.Z, li, Z.Q, Jiang, Y, Pan, X.J, Wu, J.P, Cristofori-Armstrong, B, Smith, J.J, Chin, Y.K.Y, Lei, J.L, Zhou, Q, King, G.F, Yan, N. | | 登録日 | 2018-07-11 | | 公開日 | 2018-08-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for the modulation of voltage-gated sodium channels by animal toxins.
Science, 362, 2018
|
|
6A91
 
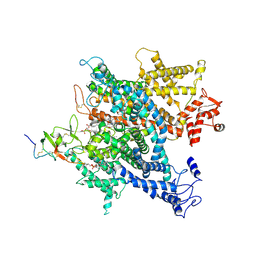 | | Complex of voltage-gated sodium channel NavPaS from American cockroach Periplaneta americana bound with saxitoxin and Dc1a | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shen, H.Z, li, Z.Q, Jiang, Y, Pan, X.J, Wu, J.P, Cristofori-Armstrong, B, Smith, J.J, Chin, Y.K.Y, Lei, J.L, Zhou, Q, King, G.F, Yan, N. | | 登録日 | 2018-07-11 | | 公開日 | 2018-08-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for the modulation of voltage-gated sodium channels by animal toxins.
Science, 362, 2018
|
|
6A95
 
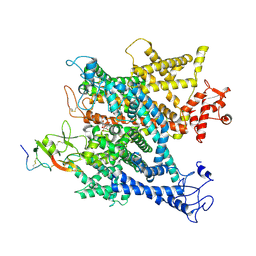 | | Complex of voltage-gated sodium channel NavPaS from American cockroach Periplaneta americana bound with tetrodotoxin and Dc1a | | 分子名称: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shen, H.Z, li, Z.Q, Jiang, Y, Pan, X.J, Wu, J.P, Cristofori-Armstrong, B, Smith, J.J, Chin, Y.K.Y, Lei, J.L, Zhou, Q, King, G.F, Yan, N. | | 登録日 | 2018-07-11 | | 公開日 | 2018-08-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural basis for the modulation of voltage-gated sodium channels by animal toxins.
Science, 362, 2018
|
|
8HLT
 
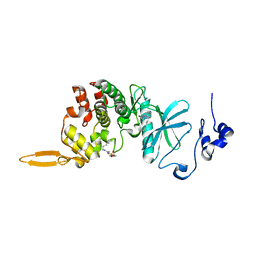 | | The co-crystal structure of DYRK2 with YK-2-99B | | 分子名称: | (6-{[(4P)-4-(1,3-benzothiazol-5-yl)-5-fluoropyrimidin-2-yl]amino}pyridin-3-yl)(piperazin-1-yl)methanone, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | 著者 | Shen, H.T, Xiao, Y.B, Yuan, K, Yang, P, Li, Q.N. | | 登録日 | 2022-12-01 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of Potent DYRK2 Inhibitors with High Selectivity, Great Solubility, and Excellent Safety Properties for the Treatment of Prostate Cancer.
J.Med.Chem., 66, 2023
|
|
5MES
 
 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 29 | | 分子名称: | (5~{R},13~{S},17~{S})-5-[[4-chloranyl-3-(2-phenylethyl)phenyl]methyl]-13-[(4-chlorophenyl)methyl]-8-methyl-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20(25),21,23-triene-3,7,15,26-tetrone, Heavy Chain, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | 著者 | Hargreaves, D. | | 登録日 | 2016-11-16 | | 公開日 | 2017-01-18 | | 最終更新日 | 2017-03-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5MEV
 
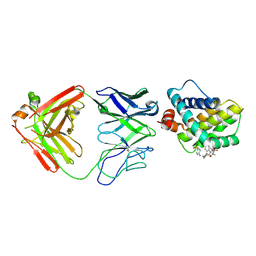 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 21 | | 分子名称: | (5~{R},13~{S},17~{S})-5-[(3,4-dichlorophenyl)methyl]-8-methyl-13-[(4-methylsulfonylphenyl)methyl]-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20,22,24-triene-3,7,15,26-tetrone, Fab Heavy Chain, Fab Light Chain, ... | | 著者 | Hargreaves, D. | | 登録日 | 2016-11-16 | | 公開日 | 2017-01-18 | | 最終更新日 | 2017-03-08 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Acs Med.Chem.Lett., 8, 2017
|
|
2G4B
 
 | |
4WVT
 
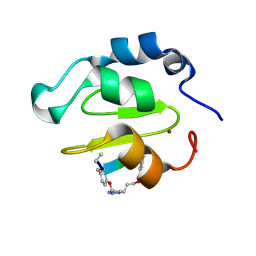 | | Crystal structure of XIAP-BIR2 domain complexed with ligand bound | | 分子名称: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, ZINC ION | | 著者 | Pokross, M.E. | | 登録日 | 2014-11-07 | | 公開日 | 2015-03-04 | | 最終更新日 | 2015-04-08 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
4WVU
 
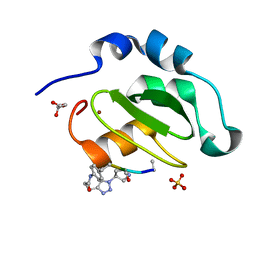 | | CRYSTAL STRUCTURE OF XIAP-BIR2 DOMAIN COMPLEXED WITH LIGAND BOUND | | 分子名称: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, GLYCEROL, ... | | 著者 | Pokross, M.E. | | 登録日 | 2014-11-07 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
4WVS
 
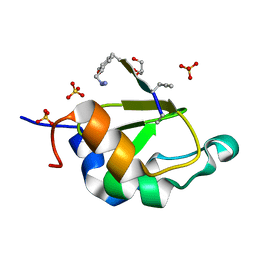 | | Crystal structure of XIAP-BIR2 domain complexed with (S)-3-(4-methoxyphenyl)-2-((S)-2-((S)-1-((S)-2-((S)-2-(methylamino)propanamido)pent-4-ynoyl)pyrrolidine-2-carboxamido)-3-phenylpropanamido)propanoic acid | | 分子名称: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, GLYCEROL, ... | | 著者 | Pokross, M.E. | | 登録日 | 2014-11-07 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
5KU9
 
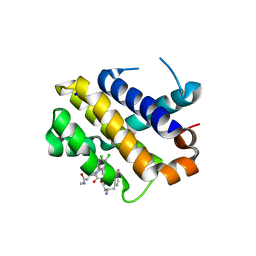 | | Crystal structure of MCL1 with compound 1 | | 分子名称: | (3~{S})-3-azanyl-4-(4-bromophenyl)-~{N}-[(3~{S})-1-[2-[[(2~{R})-1-(3,4-dichlorophenyl)-4-(methylamino)-4-oxidanylidene-butan-2-yl]amino]-2-oxidanylidene-ethyl]-2-oxidanylidene-4,5-dihydro-3~{H}-1-benzazepin-3-yl]butanamide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION | | 著者 | Ferguson, A.D. | | 登録日 | 2016-07-13 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
6XOQ
 
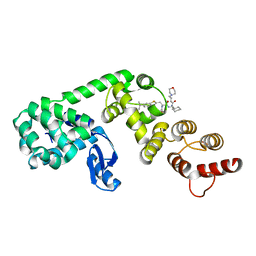 | | DCN1 covalently bound to inhibitor 4 | | 分子名称: | (2E)-N-[(2S)-2-cyclohexyl-2-({N-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alanyl}amino)ethyl]-4-(morpholin-4-yl)but-2-enamide, Lysozyme, DCN1-like protein 1 chimera | | 著者 | Stuckey, J.A. | | 登録日 | 2020-07-07 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
6XOM
 
 | | DCN1 bound to 8 | | 分子名称: | (2R)-N-[(2S)-2-cyclohexyl-2-({N-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alanyl}amino)ethyl]-2-methyl-4-(morpholin-4-yl)butanamide, 1,2-ETHANEDIOL, Lysozyme, ... | | 著者 | Stuckey, J.A. | | 登録日 | 2020-07-07 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity.
Nat Commun, 12, 2021
|
|
