3OER
 
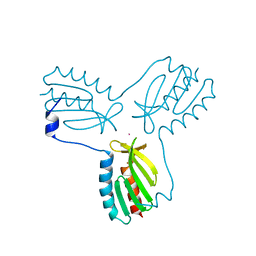 | | Crystal structure of trimeric frataxin from the yeast saccharomyces cerevisiae, complexed with cobalt | | 分子名称: | COBALT (II) ION, Frataxin homolog, mitochondrial | | 著者 | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | 登録日 | 2010-08-13 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
4N3P
 
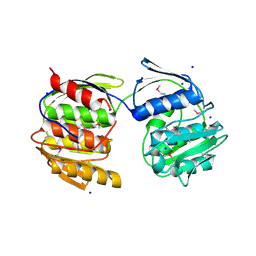 | | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396 | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Kogan, S, Maglaqui, M, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-10-07 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396
To be Published
|
|
1S1Y
 
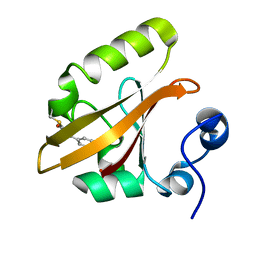 | | Photoactivated chromophore conformation in Photoactive Yellow Protein (E46Q mutant) from 10 microseconds to 3 milliseconds | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Anderson, S, Srajer, V, Pahl, R, Rajagopal, S, Schotte, F, Anfinrud, P, Wulff, M, Moffat, K. | | 登録日 | 2004-01-07 | | 公開日 | 2004-06-15 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Chromophore conformation and the evolution of tertiary structural changes in photoactive yellow protein
Structure, 12, 2004
|
|
1S1Z
 
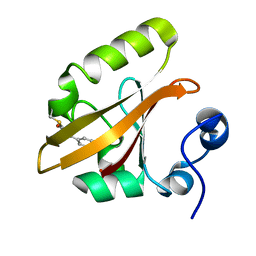 | | Photoactivated chromophore conformation in Photoactive Yellow Protein (E46Q mutant) from 10 to 500 nanoseconds | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive Yellow Protein | | 著者 | Anderson, S, Srajer, V, Pahl, R, Rajagopal, S, Schotte, F, Anfinrud, P, Wulff, M, Moffat, K. | | 登録日 | 2004-01-07 | | 公開日 | 2004-06-15 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Chromophore conformation and the evolution of tertiary structural changes in photoactive yellow protein
Structure, 12, 2004
|
|
1S4R
 
 | | Structure of a reaction intermediate in the photocycle of PYP extracted by a SVD-driven analysis | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Schmidt, M, Pahl, R, Srajer, V, Anderson, S, Ren, Z, Ihee, H, Rajagopal, S, Moffat, K. | | 登録日 | 2004-01-17 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Protein kinetics: Structures of intermediates and reaction mechanism from time-resolved x-ray data
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TS8
 
 | | Structure of the pR cis planar intermediate from time-resolved Laue crystallography | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Ihee, H, Rajagopal, S, Srajer, V, Pahl, R, Anderson, S, Schmidt, M, Schotte, F, Anfinrud, P.A, Wulff, M, Moffat, K. | | 登録日 | 2004-06-21 | | 公開日 | 2005-07-05 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1TS7
 
 | | Structure of the pR cis wobble and pR E46Q intermediates from time-resolved Laue crystallography | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Ihee, H, Rajagopal, S, Srajer, V, Pahl, R, Anderson, S, Schmidt, M, Schotte, F, Anfinrud, P.A, Wulff, M, Moffat, K. | | 登録日 | 2004-06-21 | | 公開日 | 2005-07-05 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4E3R
 
 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | 分子名称: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | 著者 | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | 登録日 | 2012-03-10 | | 公開日 | 2012-10-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4GVV
 
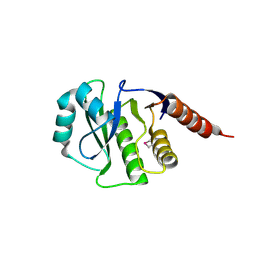 | | Crystal Structure of de novo design serine hydrolase OSH55.27, Northeast Structural Genomics Consortium (NESG) Target OR246 | | 分子名称: | De novo design serine hydrolase | | 著者 | Kuzin, A, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-08-31 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.895 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target OR246
To be Published
|
|
4GVW
 
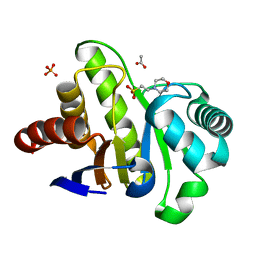 | | Three-dimensional structure of the de novo designed serine hydrolase 2bfq_3, Northeast Structural Genomics Consortium (NESG) Target OR248 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, De novo designed serine hydrolase, ... | | 著者 | Kuzin, A, Lew, S, Seetharaman, J, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-08-31 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.113 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target OR248
To be Published
|
|
1TS0
 
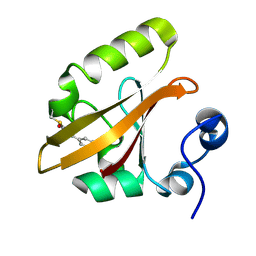 | | Structure of the pB1 intermediate from time-resolved Laue crystallography | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Ihee, H, Rajagopal, S, Srajer, V, Pahl, R, Anderson, S, Schmidt, M, Schotte, F, Anfinrud, P.A, Wulff, M, Moffat, K. | | 登録日 | 2004-06-21 | | 公開日 | 2005-07-05 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1TS6
 
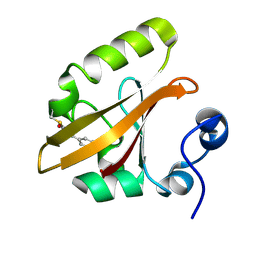 | | Structure of the pB2 intermediate from time-resolved Laue crystallography | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Ihee, H, Rajagopal, S, Srajer, V, Pahl, R, Anderson, S, Schmidt, M, Schotte, F, Anfinrud, P.A, Wulff, M, Moffat, K. | | 登録日 | 2004-06-21 | | 公開日 | 2005-07-05 | | 最終更新日 | 2021-07-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4E3Q
 
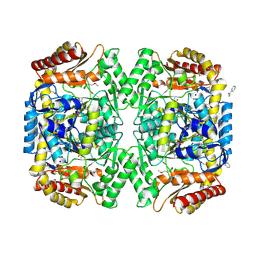 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | 著者 | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | 登録日 | 2012-03-10 | | 公開日 | 2012-10-10 | | 最終更新日 | 2013-01-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4KX6
 
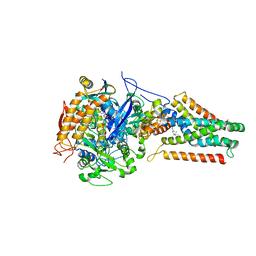 | | Plasticity of the quinone-binding site of the complex II homolog quinol:fumarate reductase | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Singh, P.K, Sarwar, M, Maklashina, E, Kotlyar, V, Rajagukguk, S, Tomasiak, T.M, Cecchini, G, Iverson, T.M. | | 登録日 | 2013-05-24 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Plasticity of the Quinone-binding Site of the Complex II Homolog Quinol:Fumarate Reductase.
J.Biol.Chem., 288, 2013
|
|
4LNV
 
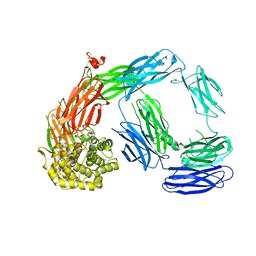 | | Crystal Structure of TEP1s | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thioester-containing protein I | | 著者 | Le, B.V, Williams, M, Logarajah, S, Baxter, R.H.G. | | 登録日 | 2013-07-12 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Molecular basis for genetic resistance of Anopheles gambiae to Plasmodium: structural analysis of TEP1 susceptible and resistant alleles.
Plos Pathog., 8, 2012
|
|
3F72
 
 | | Crystal Structure of the Staphylococcus aureus pI258 CadC Metal Binding Site 2 Mutant | | 分子名称: | Cadmium efflux system accessory protein, SODIUM ION | | 著者 | Kandegedara, A, Thiyagarajan, S, Kondapalli, K.C, Stemmler, T.L, Rosen, B.P. | | 登録日 | 2008-11-07 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Role of bound Zn(II) in the CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor.
J.Biol.Chem., 284, 2009
|
|
4G35
 
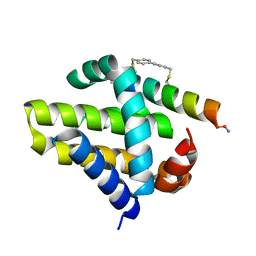 | | Mcl-1 in complex with a biphenyl cross-linked Noxa peptide. | | 分子名称: | 4,4'-bis(bromomethyl)biphenyl, Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa BH3 peptide (cysteine-mediated cross-linked) | | 著者 | Drake, E, Edwardraja, S, Lin, Q, Gulick, A.M. | | 登録日 | 2012-07-13 | | 公開日 | 2012-12-05 | | 最終更新日 | 2013-06-26 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors.
J.Am.Chem.Soc., 134, 2012
|
|
4D94
 
 | | Crystal Structure of TEP1r | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Le, B.V, Williams, M, Logarajah, S, Baxter, R.H.G. | | 登録日 | 2012-01-11 | | 公開日 | 2012-10-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Basis for Genetic Resistance of Anopheles gambiae to Plasmodium: Structural Analysis of TEP1 Susceptible and Resistant Alleles.
Plos Pathog., 8, 2012
|
|
2HTO
 
 | | Ruthenium hexammine ion interactions with Z-DNA | | 分子名称: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DCP*DGP*DCP*DG)-3'), RUTHENIUM (III) HEXAAMINE ION | | 著者 | Bharanidharan, D, Thiyagarajan, S, Gautham, N. | | 登録日 | 2006-07-26 | | 公開日 | 2006-08-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Hexammineruthenium(III) ion interactions with Z-DNA
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2HTT
 
 | | Ruthenium Hexammine ion interactions with Z-DNA | | 分子名称: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DTP*DG)-3'), ... | | 著者 | Bharanidharan, D, Thiyagarajan, S, Gautham, N. | | 登録日 | 2006-07-26 | | 公開日 | 2006-08-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Hexammineruthenium(III) ion interactions with Z-DNA
Acta Crystallogr.,Sect.F, 63, 2007
|
|
4QVF
 
 | |
4QVE
 
 | |
5Y41
 
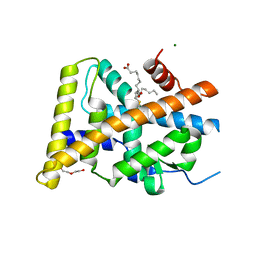 | | Crystal Structure of LIGAND-BOUND NURR1-LBD | | 分子名称: | (13E,15S)-15-hydroxy-9-oxoprosta-10,13-dien-1-oic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sreekanth, R, Lescar, J, Yoon, H.S. | | 登録日 | 2017-07-31 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function.
Nat.Chem.Biol., 2020
|
|
6P69
 
 | |
4HF2
 
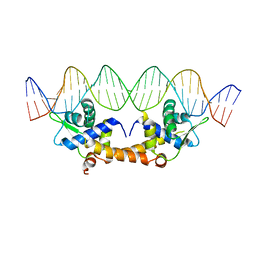 | |
