6MCR
 
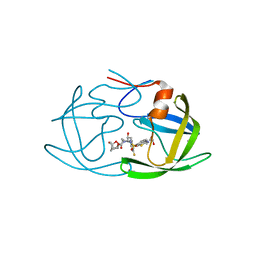 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-001 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | 著者 | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2018-09-02 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
9B2H
 
 | | HIV-1 wild type protease with GRL-072-17A, a substituted tetrahydrofuran derivative based on Darunavir as P2 group | | 分子名称: | 2,5:6,9-dianhydro-1,3,7,8-tetradeoxy-4-O-({(2S,3R)-3-hydroxy-4-[(4-methoxybenzene-1-sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamoyl)-L-gluco-nonitol, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | 登録日 | 2024-03-15 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Design of substituted tetrahydrofuran derivatives for HIV-1 protease inhibitors: synthesis, biological evaluation, and X-ray structural studies.
Org.Biomol.Chem., 2024
|
|
5DGW
 
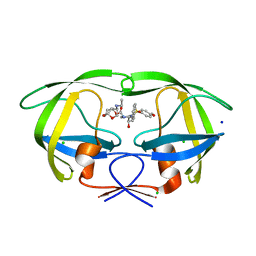 | | Crystal Structure of HIV-1 Protease Inhibitor GRL-105-11A Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand | | 分子名称: | (3R,3aS,4S,7aS)-3-(ethylamino)hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Pol protein, ... | | 著者 | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | 登録日 | 2015-08-28 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
2IEN
 
 | | Crystal structure analysis of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | 登録日 | 2006-09-19 | | 公開日 | 2006-10-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
6CDJ
 
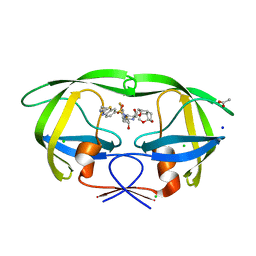 | | HIV-1 wild type protease with GRL-03314A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | 分子名称: | (2aS,4R,4aS,7aS,7bS)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2018-02-08 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
5UOV
 
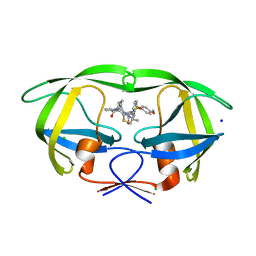 | | HIV-1 wild Type protease with GRL-1118A , an isophthalamide-derived P2-P3 ligand with the sulfonamide isostere as the P2' group | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-02-01 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
5UPZ
 
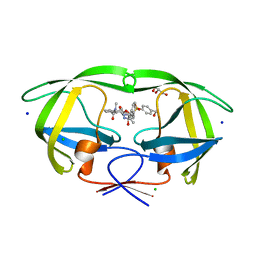 | | HIV-1 wild Type protease with GRL-0518A , an isophthalamide-derived P2-P3 ligand with the para-hydoxymethyl sulfonamide isostere as the P2' group | | 分子名称: | CHLORIDE ION, GLYCEROL, N~3~-{(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}-N~1~-methyl-N~1~-[(4-methyl-1,3-oxazol-2-yl)methyl]benzene-1,3-dicarboxamide, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-02-05 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
2HB3
 
 | | Wild-type HIV-1 Protease in complex with potent inhibitor GRL06579 | | 分子名称: | (3AS,5R,6AR)-HEXAHYDRO-2H-CYCLOPENTA[B]FURAN-5-YL (2S,3S)-3-HYDROXY-4-(4-(HYDROXYMETHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Kovalevsky, A.Y, Weber, I.T. | | 登録日 | 2006-06-13 | | 公開日 | 2006-08-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structure-Based Design of Novel HIV-1 Protease Inhibitors To Combat Drug Resistance.
J.Med.Chem., 49, 2006
|
|
6IKA
 
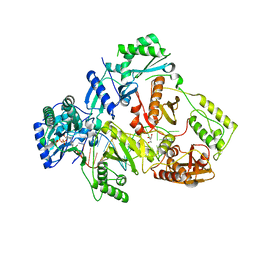 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:entecavir-triphosphate ternary complex | | 分子名称: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6IK9
 
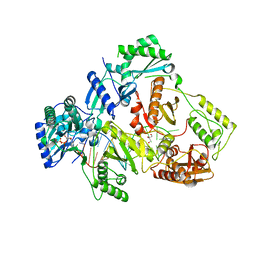 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | 著者 | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | 登録日 | 2018-10-15 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.435 Å) | | 主引用文献 | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
5ULT
 
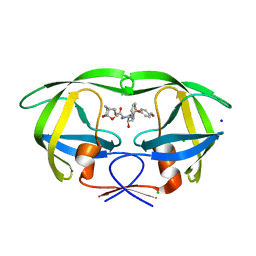 | | HIV-1 wild Type protease with GRL-100-13A (a Crown-like Oxotricyclic Core as the P2-Ligand with the sulfonamide isostere as the P2' group) | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-01-25 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Design and Development of Highly Potent HIV-1 Protease Inhibitors with a Crown-Like Oxotricyclic Core as the P2-Ligand To Combat Multidrug-Resistant HIV Variants.
J. Med. Chem., 60, 2017
|
|
5DGU
 
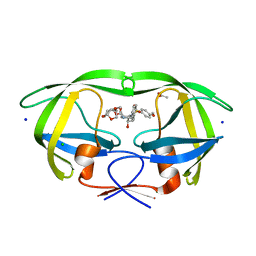 | | Crystal Structure of HIV-1 Protease Inhibitors Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand GRL-004-11A | | 分子名称: | (3R,3aR,4S,7aS)-3-methoxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | 登録日 | 2015-08-28 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
8FUJ
 
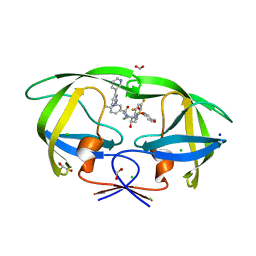 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | 分子名称: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | 登録日 | 2023-01-17 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
6DV0
 
 | | HIV-1 wild type protease with GRL-02815A, a thiochroman heterocycle with (S)-Boc-amine functionality as the P2 ligand | | 分子名称: | CHLORIDE ION, GLYCEROL, Protease, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2018-06-22 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors incorporating aminothiochromane and aminotetrahydronaphthalene carboxamide derivatives as the P2 ligands.
Eur J Med Chem, 160, 2018
|
|
6DV4
 
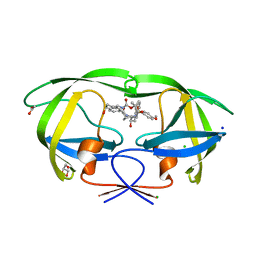 | | HIV-1 wild type protease with GRL-04315A, a tetrahydronaphthalene carboxamide with (R)-Boc-amine and (S)-hydroxyl functionalities as the P2 ligand | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2018-06-22 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors incorporating aminothiochromane and aminotetrahydronaphthalene carboxamide derivatives as the P2 ligands.
Eur J Med Chem, 160, 2018
|
|
4JLH
 
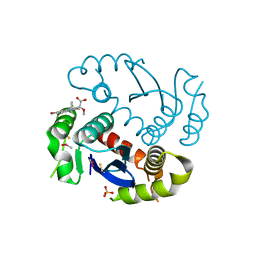 | |
4KB9
 
 | | Crystal structure of wild-type HIV-1 protease with novel tricyclic P2-ligands GRL-0739A | | 分子名称: | (3aR,3bR,4S,7aR,8aS)-decahydrofuro[2,3-b][1]benzofuran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2013-04-23 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Highly Potent HIV-1 Protease Inhibitors with Novel Tricyclic P2 Ligands: Design, Synthesis, and Protein-Ligand X-ray Studies.
J.Med.Chem., 56, 2013
|
|
4U8W
 
 | | HIV-1 wild Type protease with GRL-050-10A (a Gem-difluoro-bis-Tetrahydrofuran as P2-Ligand) | | 分子名称: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2014-08-05 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Design of gem-Difluoro-bis-Tetrahydrofuran as P2 Ligand for HIV-1 Protease Inhibitors to Improve Brain Penetration: Synthesis, X-ray Studies, and Biological Evaluation.
Chemmedchem, 10, 2015
|
|
1HPX
 
 | | HIV-1 PROTEASE COMPLEXED WITH THE INHIBITOR KNI-272 | | 分子名称: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, HIV-1 PROTEASE | | 著者 | Bhat, T.N, Erickson, J.W. | | 登録日 | 1995-05-18 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of HIV-1 protease with KNI-272, a tight-binding transition-state analog containing allophenylnorstatine.
Structure, 3, 1995
|
|
5XN2
 
 | | HIV-1 reverse transcriptase Q151M:DNA:dGTP ternary complex | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 38-MER DNA aptamer, GLYCEROL, ... | | 著者 | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | 登録日 | 2017-05-17 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.381 Å) | | 主引用文献 | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN0
 
 | | HIV-1 reverse transcriptase Q151M:DNA binary complex | | 分子名称: | 38-MER DNA aptamer, GLYCEROL, Pol protein, ... | | 著者 | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | 登録日 | 2017-05-17 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.596 Å) | | 主引用文献 | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
5XN1
 
 | | HIV-1 reverse transcriptase Q151M:DNA:entecavir-triphosphate ternary complex | | 分子名称: | 38-MER DNA aptamer, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Yasutake, Y, Tamura, N, Hayashi, H, Maeda, K. | | 登録日 | 2017-05-17 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.446 Å) | | 主引用文献 | HIV-1 with HBV-associated Q151M substitution in RT becomes highly susceptible to entecavir: structural insights into HBV-RT inhibition by entecavir.
Sci Rep, 8, 2018
|
|
7RC0
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-091-20 | | 分子名称: | 3C-like proteinase, 5-chloro-4-methylpyridin-3-yl 1H-indole-4-carboxylate, SODIUM ION | | 著者 | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-06 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7RC1
 
 | | X-ray Structure of SARS-CoV main protease covalently modified by compound GRL-0686 | | 分子名称: | 3C-like proteinase, 5-chloropyridin-3-yl 1-(3-nitrobenzene-1-sulfonyl)-1H-indole-5-carboxylate, DIMETHYL SULFOXIDE | | 著者 | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-07 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7RBZ
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-017-20 | | 分子名称: | 3C-like proteinase, 5-chloropyridin-3-yl 2,3-dihydro-1H-indole-4-carboxylate | | 著者 | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-06 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
