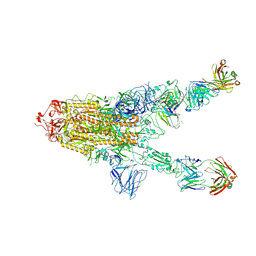
7MW5
 
 | |
8D4L
 
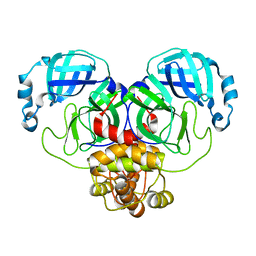 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4K
 
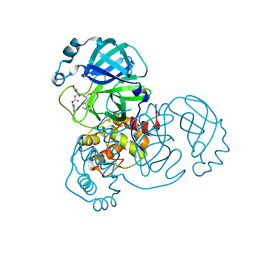 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4M
 
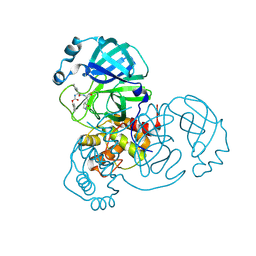 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4J
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4N
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166Q Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
2MQW
 
 | |
2MRI
 
 | |
2MR3
 
 | |
8GQ6
 
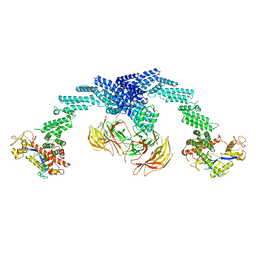 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | 登録日 | 2022-08-29 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8DTY
 
 | |
8DVV
 
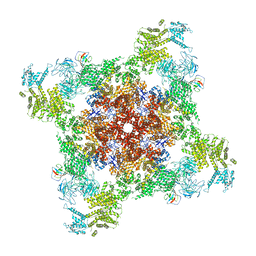 | | Recombinant mouse RyR2 triple phosphomimetic mutant S2807D/S2813D/S2030D in complex with FKBP12.6 and nanodisc under open-state conditions | | 分子名称: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | 著者 | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | 登録日 | 2022-07-29 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Recombinant mouse RyR2 triple phosphomimetic mutant S2807D/S2813D/S2030D in complex with FKBP12.6 and nanodisc under open-state conditions
To Be Published
|
|
8DTZ
 
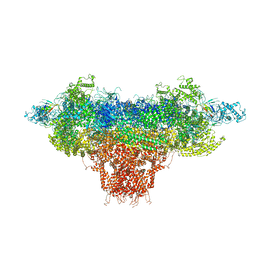 | |
3SWJ
 
 | | Crystal structure of Campylobacter jejuni ChuZ | | 分子名称: | AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein | | 著者 | Hu, Y. | | 登録日 | 2011-07-14 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.409 Å) | | 主引用文献 | Crystal structure of Campylobacter jejuni ChuZ: a split-barrel family heme oxygenase with a novel heme-binding mode.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
2IPA
 
 | | solution structure of Trx-ArsC complex | | 分子名称: | Protein arsC, Thioredoxin | | 著者 | Jin, C, Hu, Y, Li, Y, Zhang, X. | | 登録日 | 2006-10-12 | | 公開日 | 2007-02-13 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis.
J.Biol.Chem., 282, 2007
|
|
6VXT
 
 | | Activated Nitrogenase MoFe-protein from Azotobacter vinelandii | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Kang, W, Hu, Y, Ribbe, M.W. | | 登録日 | 2020-02-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
5B7J
 
 | | Structure model of Sap1-DNA complex | | 分子名称: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*GP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*AP*TP*AP*TP*T)-3'), Switch-activating protein 1 | | 著者 | Jin, C, Hu, Y, Ding, J, Zhang, Y. | | 登録日 | 2016-06-07 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe
J. Biol. Chem., 292, 2017
|
|
4D8M
 
 | | Crystal structure of Bacillus thuringiensis Cry5B nematocidal toxin | | 分子名称: | Pesticidal crystal protein cry5Ba | | 著者 | Fan, H, Hu, Y, Aroian, R.V, Ghosh, P, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2012-01-10 | | 公開日 | 2012-12-19 | | 最終更新日 | 2013-02-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and Glycolipid Binding Properties of the Nematicidal Protein Cry5B.
Biochemistry, 51, 2012
|
|
1XVA
 
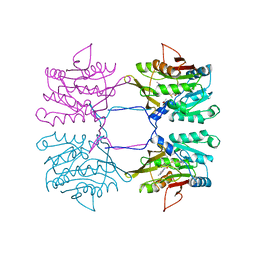 | | METHYLTRANSFERASE | | 分子名称: | ACETATE ION, GLYCINE N-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | 著者 | Fu, Z, Hu, Y, Konishi, K, Takata, Y, Ogawa, H, Gomi, T, Fujioka, M, Takusagawa, F. | | 登録日 | 1996-07-20 | | 公開日 | 1997-01-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glycine N-methyltransferase from rat liver.
Biochemistry, 35, 1996
|
|
4KQZ
 
 | | structure of the receptor binding domain (RBD) of MERS-CoV spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Bao, J, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2013-05-15 | | 公開日 | 2013-07-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.514 Å) | | 主引用文献 | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
1LI4
 
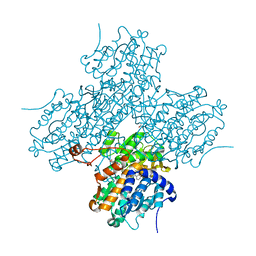 | | Human S-adenosylhomocysteine hydrolase complexed with neplanocin | | 分子名称: | 3-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-CYCLOPENTANE-1,2-DIOL, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Yang, X, Hu, Y, Yin, D.H, Turner, M.A, Wang, M, Borchardt, R.T, Howell, P.L, Kuczera, K, Schowen, R.L. | | 登録日 | 2002-04-17 | | 公開日 | 2003-05-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Catalytic strategy of S-adenosyl-L-homocysteine hydrolase: Transition-state
stabilization and the avoidance of abortive reactions
Biochemistry, 42, 2003
|
|
4KR0
 
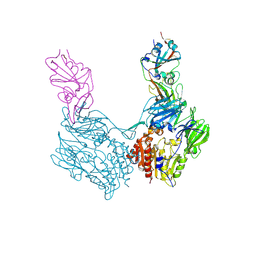 | | Complex structure of MERS-CoV spike RBD bound to CD26 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | 著者 | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2013-05-15 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
3LKX
 
 | | Human nac dimerization domain | | 分子名称: | Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | 著者 | Liu, Y, Hu, Y, Li, X, Niu, L, Teng, M. | | 登録日 | 2010-01-28 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit
Biochemistry, 49, 2010
|
|
2MYP
 
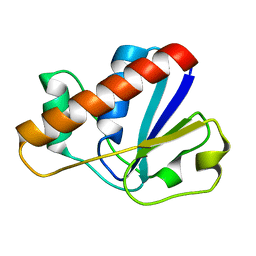 | |
2MYT
 
 | |
