2BFX
 
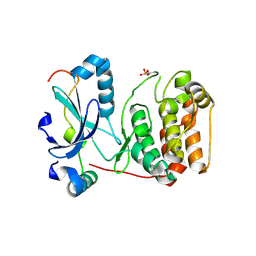 | | Mechanism of Aurora-B activation by INCENP and inhibition by Hesperadin. | | 分子名称: | AURORA KINASE B-A, INNER CENTROMERE PROTEIN A | | 著者 | Sessa, F, Mapelli, M, Ciferri, C, Tarricone, C, Areces, L.B, Schneider, T.R, Stukenberg, P.T, Musacchio, A. | | 登録日 | 2004-12-15 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mechanism of Aurora B Activation by Incenp and Inhibition by Hesperadin
Mol.Cell, 18, 2005
|
|
2C7M
 
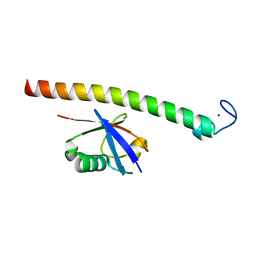 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | 分子名称: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | 著者 | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | 登録日 | 2005-11-25 | | 公開日 | 2006-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin.
Cell, 124, 2006
|
|
2C7N
 
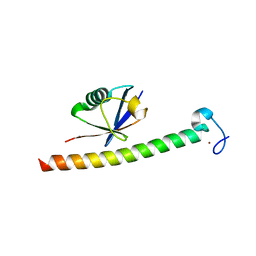 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | 分子名称: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | 著者 | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | 登録日 | 2005-11-25 | | 公開日 | 2006-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Ubiquitin Binding Domains of Rabex-5 Reveals Two Modes of Interaction with Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
4C1O
 
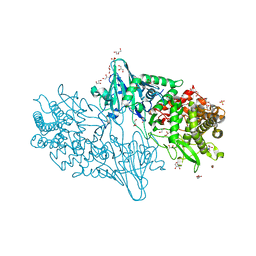 | | Geobacillus thermoglucosidasius GH family 52 xylosidase | | 分子名称: | 1,2-ETHANEDIOL, BETA-XYLOSIDASE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Espina, G, Eley, K, Schneider, T.R, Crennell, S.J, Danson, M.J. | | 登録日 | 2013-08-13 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A Novel Beta-Xylosidase Structure from Geobacillus Thermoglucosidasius: The First Crystal Structure of a Glycoside Hydrolase Family Gh52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BLG
 
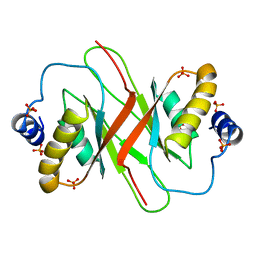 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | 分子名称: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | 著者 | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | 登録日 | 2013-05-02 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
4C1P
 
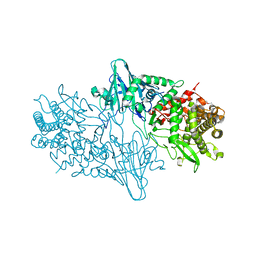 | | Geobacillus thermoglucosidasius GH family 52 xylosidase | | 分子名称: | BETA-XYLOSIDASE, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Espina, G, Eley, K, Schneider, T.R, Crennell, S.J, Danson, M.J. | | 登録日 | 2013-08-13 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.634 Å) | | 主引用文献 | A Novel Beta-Xylosidase Structure from Geobacillus Thermoglucosidasius: The First Crystal Structure of a Glycoside Hydrolase Family Gh52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1AR2
 
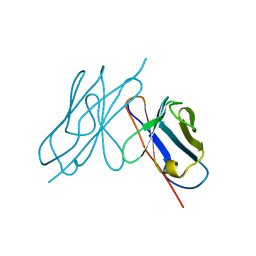 | | DISULFIDE-FREE IMMUNOGLOBULIN FRAGMENT | | 分子名称: | REI | | 著者 | Uson, I, Bes, M.T, Sheldrick, G.M, Schneider, T.R, Hartsch, T, Fritz, H.-J. | | 登録日 | 1997-08-08 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | X-ray crystallography reveals stringent conservation of protein fold after removal of the only disulfide bridge from a stabilized immunoglobulin variable domain.
Structure Fold.Des., 2, 1997
|
|
1BWW
 
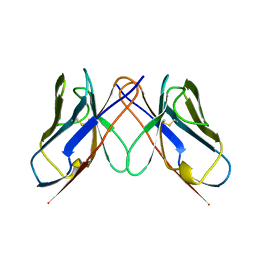 | | BENCE-JONES IMMUNOGLOBULIN REI VARIABLE PORTION, T39K MUTANT | | 分子名称: | PROTEIN (IG KAPPA CHAIN V-I REGION REI) | | 著者 | Uson, I, Pohl, E, Schneider, T.R, Dauter, Z, Schmidt, A, Fritz, H.J, Sheldrick, G.M. | | 登録日 | 1998-09-29 | | 公開日 | 1998-10-07 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 1.7 A structure of the stabilized REIv mutant T39K. Application of local NCS restraints.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6ZBV
 
 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor and sybody Sb_GlyT1#7 | | 分子名称: | Sodium- and chloride-dependent glycine transporter 1,Sodium- and chloride-dependent glycine transporter 1, Sybody Sb_GlyT1#7, [5-fluoranyl-6-(oxan-4-yloxy)-1,3-dihydroisoindol-2-yl]-[5-methylsulfonyl-2-[2,2,3,3,3-pentakis(fluoranyl)propoxy]phenyl]methanone | | 著者 | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | 登録日 | 2020-06-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
6ZPL
 
 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | 分子名称: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | 著者 | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | 登録日 | 2020-07-08 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.945 Å) | | 主引用文献 | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
7AF2
 
 | | Salmonella typhimurium neuraminidase mutant (D62G) | | 分子名称: | GLYCEROL, PHOSPHATE ION, Sialidase | | 著者 | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vimr, E.R, Garman, E.F. | | 登録日 | 2020-09-19 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.792 Å) | | 主引用文献 | Salmonella typhimurium neuraminidase mutant (D62G)
To Be Published
|
|
7AEY
 
 | | Salmonella typhimurium neuraminidase in complex with isocarba-DANA. | | 分子名称: | (3~{S},4~{S},5~{R})-4-acetamido-3-oxidanyl-5-[(1~{S},2~{R})-1,2,3-tris(oxidanyl)propyl]cyclohexane-1-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vasella, A.T, Vimr, E.R, Vorwerk, S, Garman, E.F. | | 登録日 | 2020-09-18 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.919 Å) | | 主引用文献 | Salmonella typhimurium neuraminidase in complex with isocarba-DANA.
To Be Published
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | 分子名称: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2021-02-05 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | 分子名称: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-04-14 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | 分子名称: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-04-28 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8C53
 
 | | Trypanosoma brucei IMP dehydrogenase (ori) crystallized in High Five cells reveals native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo; CrystFEL processing | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | 著者 | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Bourenkov, G, Schneider, T, Redecke, L. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C51
 
 | | Trypanosoma brucei IMP dehydrogenase (cyto) crystallized in High Five cells revealing native ligands ATP, GDP and phosphate. Diffraction data collection at 100 K in cellulo | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | 著者 | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
8C5K
 
 | | HEX-1 (in cellulo, in situ) crystallized and diffracted in High Five cells. Growth and SX data collection at 296 K on CrystalDirect plates | | 分子名称: | Woronin body major protein | | 著者 | Lahey-Rudolph, J.M, Schoenherr, R, Boger, J, Harms, M, Kaiser, J, Nachtschatt, S, Wobbe, M, Duden, R, Koenig, P, Bourenkov, G, Schneider, T, Redecke, L. | | 登録日 | 2023-01-09 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | A streamlined approach to structure elucidation using in cellulo crystallized recombinant proteins, InCellCryst.
Nat Commun, 15, 2024
|
|
6R85
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-glutamate | | 分子名称: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 3.3,Glutamate receptor 3.3, ... | | 著者 | Alfieri, A, Pederzoli, R, Costa, A. | | 登録日 | 2019-03-31 | | 公開日 | 2020-01-01 | | 最終更新日 | 2020-01-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R8A
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-methionine | | 分子名称: | Glutamate receptor 3.3,Glutamate receptor 3.3, METHIONINE, SODIUM ION, ... | | 著者 | Alfieri, A, Pederzoli, R, Costa, A. | | 登録日 | 2019-04-01 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R88
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | 分子名称: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | 著者 | Alfieri, A, Pederzoli, R, Costa, A. | | 登録日 | 2019-04-01 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R89
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | 分子名称: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | 著者 | Alfieri, A, Pederzoli, R, Costa, A. | | 登録日 | 2019-04-01 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RNC
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Lysozyme with GlcNAc3 - 100ms diffusion time. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | 登録日 | 2019-05-08 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.799 Å) | | 主引用文献 | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6RNB
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Lysozyme with GlcNAc3 50ms diffusion time | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | 著者 | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | 登録日 | 2019-05-08 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.699 Å) | | 主引用文献 | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
6RNF
 
 | | Liquid Application Method for time-resolved Analyses (LAMA) by serial synchrotron crystallography, Xylose Isomerase 30 ms timepoint | | 分子名称: | MAGNESIUM ION, Xylose isomerase, alpha-D-glucopyranose | | 著者 | Mehrabi, P, Schulz, E.C, Miller, R.J.D. | | 登録日 | 2019-05-08 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Liquid application method for time-resolved analyses by serial synchrotron crystallography.
Nat.Methods, 16, 2019
|
|
