7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | 著者 | Wang, W.W, Wu, H, He, J.H, Yu, F. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
7DWO
 
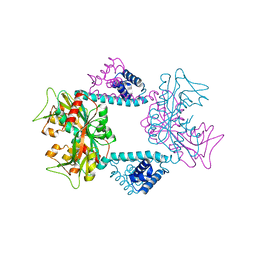 | |
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-12-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
8HQI
 
 | |
8HQH
 
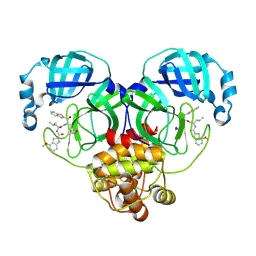 | |
4NJL
 
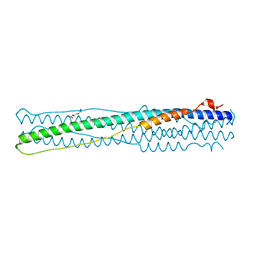 | | Crystal structure of middle east respiratory syndrome coronavirus S2 protein fusion core | | 分子名称: | S protein, TRIETHYLENE GLYCOL | | 著者 | Zhu, Y, Lu, L, Qin, L, Ye, S, Jiang, S, Zhang, R. | | 登録日 | 2013-11-10 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor.
Nat Commun, 5, 2014
|
|
8JJ2
 
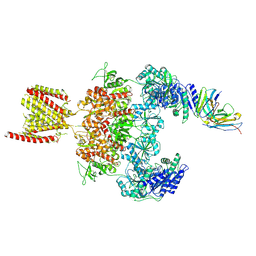 | |
8JJ1
 
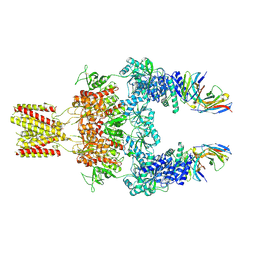 | |
8JIZ
 
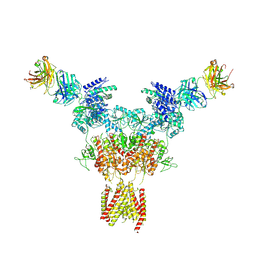 | |
8JJ0
 
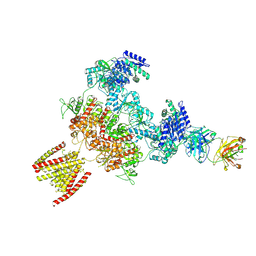 | |
7E3J
 
 | |
3HVA
 
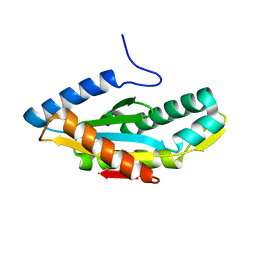 | |
3HV8
 
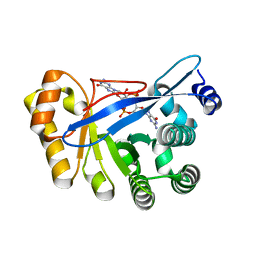 | | Crystal structure of FimX EAL domain from Pseudomonas aeruginosa bound to c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Protein FimX | | 著者 | Navarro, M.V.A.S, De, N, Bae, N, Sondermann, H. | | 登録日 | 2009-06-15 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.445 Å) | | 主引用文献 | Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX.
Structure, 17, 2009
|
|
3M45
 
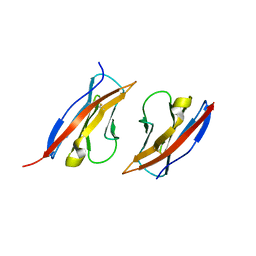 | | Crystal structure of Ig1 domain of mouse SynCAM 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 2 | | 著者 | Yue, L, Modis, Y. | | 登録日 | 2010-03-10 | | 公開日 | 2010-09-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | N-glycosylation at the SynCAM (synaptic cell adhesion molecule) immunoglobulin interface modulates synaptic adhesion.
J.Biol.Chem., 285, 2010
|
|
7TQQ
 
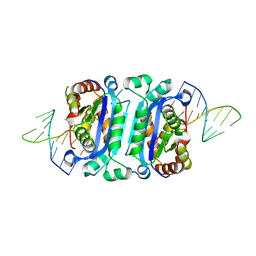 | |
7TQO
 
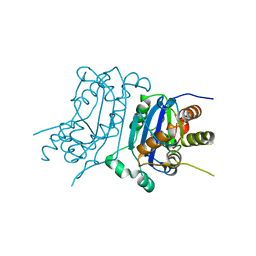 | |
7TQN
 
 | |
7TQP
 
 | |
7TTI
 
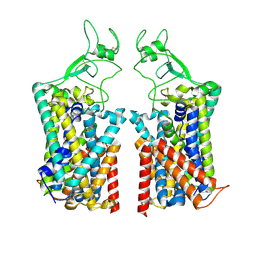 | | Human KCC1 bound with VU0463271 In an outward-open state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-cyclopropyl-N-(4-methyl-1,3-thiazol-2-yl)-2-[(6-phenylpyridazin-3-yl)sulfanyl]acetamide, Solute carrier family 12 member 4 | | 著者 | Zhao, Y.X, Cao, E.H. | | 登録日 | 2022-02-01 | | 公開日 | 2022-06-29 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TTH
 
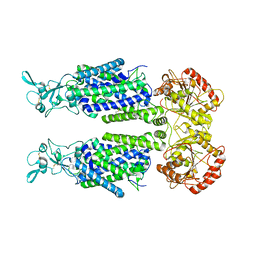 | | Human potassium-chloride cotransporter 1 in inward-open state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, POTASSIUM ION, Solute carrier family 12 member 4 | | 著者 | Zhao, Y.X, Cao, E.H. | | 登録日 | 2022-02-01 | | 公開日 | 2022-06-29 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7FIK
 
 | | The cryo-EM structure of the CR subunit from X. laevis NPC | | 分子名称: | MGC154553 protein, MGC83295 protein, MGC83926 protein, ... | | 著者 | Shi, Y, Huang, G, Zhan, X. | | 登録日 | 2021-07-31 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex.
Science, 376, 2022
|
|
7FIL
 
 | |
8ZC5
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-15 | | 最終更新日 | 2025-07-16 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC0
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC3
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-15 | | 最終更新日 | 2025-07-16 | | 実験手法 | ELECTRON MICROSCOPY (4.69 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
