1IH7
 
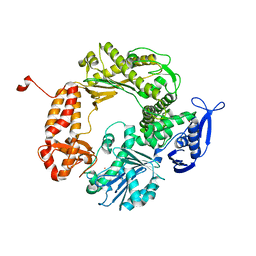 | | High-Resolution Structure of Apo RB69 DNA Polymerase | | 分子名称: | DNA POLYMERASE, GUANOSINE, POTASSIUM ION | | 著者 | Franklin, M.C, Wang, J, Steitz, T.A. | | 登録日 | 2001-04-18 | | 公開日 | 2001-06-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure of the Replicating Complex of a Pol alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
3FWE
 
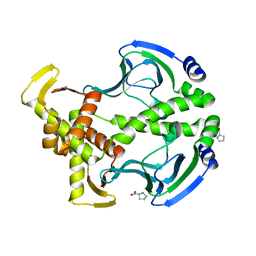 | | Crystal Structure of the Apo D138L CAP mutant | | 分子名称: | Catabolite gene activator, PROLINE | | 著者 | Sharma, H, Wang, J, Kong, J, Yu, S, Steitz, T. | | 登録日 | 2009-01-17 | | 公開日 | 2009-09-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7TOB
 
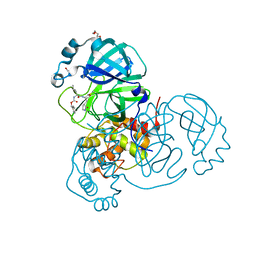 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Sacco, M.D, Wang, J, Chen, Y. | | 登録日 | 2022-01-24 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
7TQL
 
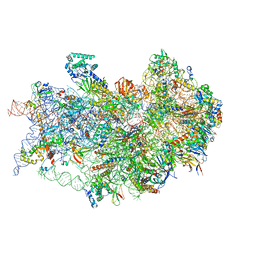 | | CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B. | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Lapointe, C.P, Grosely, R, Sokabe, M, Alvarado, C, Wang, J, Montabana, E, Villa, N, Shin, B, Dever, T, Fraser, C, Fernandez, I.S, Puglisi, J.D. | | 登録日 | 2022-01-26 | | 公開日 | 2022-04-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | eIF5B and eIF1A reorient initiator tRNA to allow ribosomal subunit joining.
Nature, 607, 2022
|
|
4EBY
 
 | | Crystal structure of the ectodomain of a receptor like kinase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | 著者 | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | 登録日 | 2012-03-25 | | 公開日 | 2012-06-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
1M4B
 
 | | Crystal Structure of Human Interleukin-2 K43C Covalently Modified at C43 with 2-[2-(2-Cyclohexyl-2-guanidino-acetylamino)-acetylamino]-N-(3-mercapto-propyl)-propionamide | | 分子名称: | 2-[2-(2-CYCLOHEXYL-2-GUANIDINO-ACETYLAMINO)-ACETYLAMINO]-N-(3-MERCAPTO-PROPYL)-PROPIONAMIDE, interleukin-2 | | 著者 | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | 登録日 | 2002-07-02 | | 公開日 | 2002-07-31 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5JPS
 
 | |
5KTJ
 
 | | Crystal structure of Pistol, a class of self-cleaving ribozyme | | 分子名称: | COBALT HEXAMMINE(III), MAGNESIUM ION, Pistol (50-MER), ... | | 著者 | Nguyen, L.A, Wang, J, Steitz, T.A. | | 登録日 | 2016-07-11 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | Crystal structure of Pistol, a class of self-cleaving ribozyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3CMZ
 
 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | 分子名称: | Beta-lactamase TEM, PHOSPHATE ION | | 著者 | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | 登録日 | 2008-03-24 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
3QNO
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dATP Opposite 3tCo | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA Polymerase, ... | | 著者 | Xia, S, Wang, M, Wang, J, Konigsberg, W.H. | | 登録日 | 2011-02-08 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Using a Fluorescent Cytosine Analogue tC(o) To Probe the Effect of the Y567 to Ala Substitution on the Preinsertion Steps of dNMP Incorporation by RB69 DNA Polymerase.
Biochemistry, 51, 2012
|
|
1EFG
 
 | | THE CRYSTAL STRUCTURE OF ELONGATION FACTOR G COMPLEXED WITH GDP, AT 2.7 ANGSTROMS RESOLUTION | | 分子名称: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | 登録日 | 1994-10-17 | | 公開日 | 1995-02-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
6IFN
 
 | | Crystal structure of Type III-A CRISPR Csm complex | | 分子名称: | MANGANESE (II) ION, RNA (32-MER), Type III-A CRISPR-associated RAMP protein Csm3, ... | | 著者 | You, L, Wang, J, Wang, Y. | | 登録日 | 2018-09-20 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
3KCU
 
 | | Structure of formate channel | | 分子名称: | 2-(6-(2-CYCLOHEXYLETHOXY)-TETRAHYDRO-4,5-DIHYDROXY-2(HYDROXYMETHYL)-2H-PYRAN-3-YLOXY)-TETRAHYDRO-6(HYDROXYMETHYL)-2H-PY RAN-3,4,5-TRIOL, Probable formate transporter 1 | | 著者 | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | 登録日 | 2009-10-22 | | 公開日 | 2009-12-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.243 Å) | | 主引用文献 | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
8TMT
 
 | | Crystal structure of KPC-44 carbapenemase in complex with vaborbactam | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, LITHIUM ION, ... | | 著者 | Sun, Z, Palzkill, T, Hu, L, Neetu, N, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | 登録日 | 2023-07-30 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
7CUN
 
 | | The structure of human Integrator-PP2A complex | | 分子名称: | Integrator complex subunit 1, Integrator complex subunit 11, Integrator complex subunit 2, ... | | 著者 | Zheng, H, Qi, Y, Liu, W, Li, J, Wang, J, Xu, Y. | | 登録日 | 2020-08-23 | | 公開日 | 2020-11-25 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Science, 370, 2020
|
|
8TN0
 
 | | Crystal structure of KPC-44 carbapenemase w/o cryoprotectant | | 分子名称: | SULFATE ION, beta-lactamase | | 著者 | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | 登録日 | 2023-07-31 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TJM
 
 | | Crystal structure of KPC-44 carbapenemase | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, beta-lactamase | | 著者 | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | 登録日 | 2023-07-23 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
8TMR
 
 | | Crystal structure of KPC-44 carbapenemase complexed with avibactam | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | 著者 | Sun, Z, Palzkill, T, Hu, L, Lin, H, Sankaran, B, Wang, J, Prasad, B. | | 登録日 | 2023-07-30 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Klebsiella pneumoniae carbapenemase variant 44 acquires ceftazidime-avibactam resistance by altering the conformation of active-site loops.
J.Biol.Chem., 300, 2023
|
|
6BT5
 
 | | Human mGlu8 Receptor complexed with L-AP4 | | 分子名称: | (2S)-2-amino-4-phosphonobutanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 8 | | 著者 | Schkeryantz, J.M, Chen, Q, Ho, J.D, Atwell, S, Zhang, A, Vargas, M.C, Wang, J, Monn, J.A, Hao, J. | | 登録日 | 2017-12-05 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Determination of L-AP4-bound human mGlu8 receptor amino terminal domain structure and the molecular basis for L-AP4's group III mGlu receptor functional potency and selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6BSZ
 
 | | Human mGlu8 Receptor complexed with glutamate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 8, ... | | 著者 | Schkeryantz, J.M, Chen, Q, Ho, J.D, Atwell, S, Zhang, A, Vargas, M.C, Wang, J, Monn, J.A, Hao, J. | | 登録日 | 2017-12-04 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Determination of L-AP4-bound human mGlu8 receptor amino terminal domain structure and the molecular basis for L-AP4's group III mGlu receptor functional potency and selectivity.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
2PQE
 
 | | Solution structure of proline-free mutant of staphylococcal nuclease | | 分子名称: | Thermonuclease | | 著者 | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | 登録日 | 2007-05-01 | | 公開日 | 2007-06-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
7LJY
 
 | | Cryo-EM structure of the B dENE construct complexed with a 28-mer poly(A) | | 分子名称: | B dENE construct, poly(A) | | 著者 | Torabi, S, Chen, Y, Zhang, K, Wang, J, DeGregorio, S, Vaidya, A, Su, Z, Pabit, S, Chiu, W, Pollack, L, Steitz, J. | | 登録日 | 2021-02-01 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Structural analyses of an RNA stability element interacting with poly(A).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4EBZ
 
 | | Crystal structure of the ectodomain of a receptor like kinase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | 著者 | Chai, J, Liu, T, Han, Z, She, J, Wang, J. | | 登録日 | 2012-03-26 | | 公開日 | 2012-06-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Chitin-induced dimerization activates a plant immune receptor.
Science, 336, 2012
|
|
1K1D
 
 | | Crystal structure of D-hydantoinase | | 分子名称: | D-hydantoinase, ZINC ION | | 著者 | Cheon, Y.H, Kim, H.S, Han, K.H, Abendroth, J, Niefind, K, Schomburg, D, Wang, J, Kim, Y. | | 登録日 | 2001-09-25 | | 公開日 | 2002-08-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Crystal structure of D-hydantoinase from Bacillus stearothermophilus: insight into the stereochemistry of enantioselectivity.
Biochemistry, 41, 2002
|
|
2EX3
 
 | | Bacteriophage phi29 DNA polymerase bound to terminal protein | | 分子名称: | DNA polymerase, DNA terminal protein, LEAD (II) ION | | 著者 | Kamtekar, S, Berman, A.J, Wang, J, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | 登録日 | 2005-11-07 | | 公開日 | 2006-03-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition
Embo J., 25, 2006
|
|
