5CFW
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7STI
 
 | |
7STJ
 
 | |
7STK
 
 | |
7STH
 
 | |
8CZK
 
 | |
8D19
 
 | | Human LanCL1 bound to GSH | | 分子名称: | GLUTATHIONE, Glutathione S-transferase LANCL1, ZINC ION | | 著者 | Ongpipattanakul, C, Nair, S.K. | | 登録日 | 2022-05-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8CZL
 
 | |
8D0V
 
 | |
8DL5
 
 | |
8DSO
 
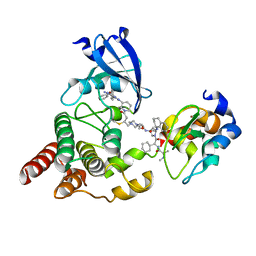 | | Structure of cIAP1, BTK and BCCov | | 分子名称: | (4S)-4-[2-(2-{4-[(2E)-4-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}-4-oxobut-2-en-1-yl]piperazin-1-yl}ethoxy)acetamido]-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]-L-prolinamide bound form, Baculoviral IAP repeat-containing protein 2, Tyrosine-protein kinase BTK, ... | | 著者 | Schiemer, J.S, Calabrese, M.F. | | 登録日 | 2022-07-22 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.334 Å) | | 主引用文献 | A covalent BTK ternary complex compatible with targeted protein degradation.
Nat Commun, 14, 2023
|
|
8DSF
 
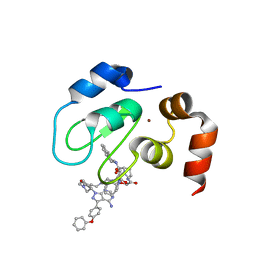 | | Structure of cIAP1 with BCCov | | 分子名称: | (4S)-4-[2-(2-{4-[(2E)-4-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}-4-oxobut-2-en-1-yl]piperazin-1-yl}ethoxy)acetamido]-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]-L-prolinamide unbound form, Baculoviral IAP repeat-containing protein 2, ZINC ION | | 著者 | Schiemer, J.S, Calabrese, M.F. | | 登録日 | 2022-07-22 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A covalent BTK ternary complex compatible with targeted protein degradation.
Nat Commun, 14, 2023
|
|
7KPY
 
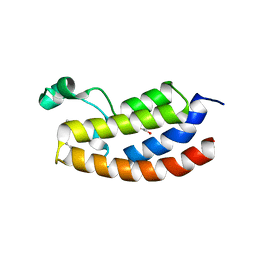 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | 分子名称: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | 著者 | Schonbrunn, E, Bikowitz, M. | | 登録日 | 2020-11-12 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8FBF
 
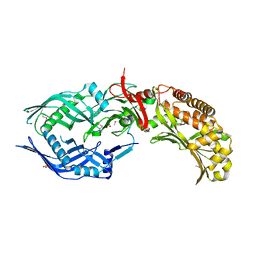 | |
7E7Y
 
 | |
7E7X
 
 | |
7E88
 
 | |
7E86
 
 | |
6BFG
 
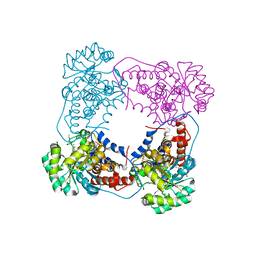 | |
5YQN
 
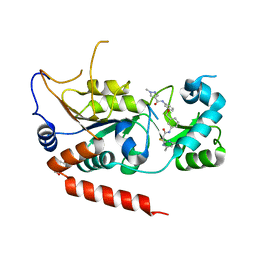 | | Crystal structure of Sirt2 in complex with selective inhibitor L55 | | 分子名称: | DI(HYDROXYETHYL)ETHER, N-[3-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ... | | 著者 | Wang, H, Yu, Y, Li, G, Chen, Q. | | 登録日 | 2017-11-07 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQO
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor L5C | | 分子名称: | N-[4-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | 著者 | Wang, H, Yu, Y, Li, G, Chen, Q. | | 登録日 | 2017-11-07 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.483 Å) | | 主引用文献 | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQL
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor A2I | | 分子名称: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[3-(phenoxymethyl)phenyl]ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | 著者 | Wang, H, Yu, Y, Li, G, Chen, Q. | | 登録日 | 2017-11-07 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
5YQM
 
 | | Crystal structure of Sirt2 in complex with selective inhibitor A29 | | 分子名称: | 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-(4-phenylsulfanylphenyl)ethanamide, BETA-MERCAPTOETHANOL, NAD-dependent protein deacetylase sirtuin-2, ... | | 著者 | Wang, H, Yu, Y, Li, G, chen, Q. | | 登録日 | 2017-11-07 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.735 Å) | | 主引用文献 | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
