6W5J
 
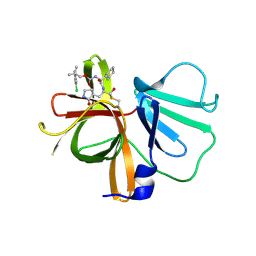 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7d | | 分子名称: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-03-13 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
5UZC
 
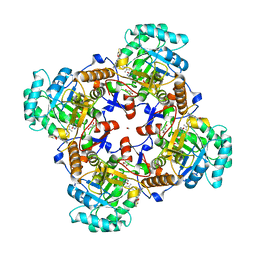 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P221 | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | 著者 | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-26 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P221
To Be Published
|
|
6UHE
 
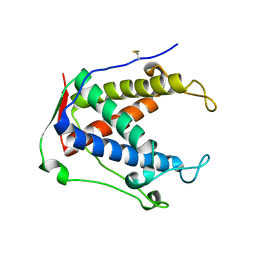 | |
6UHI
 
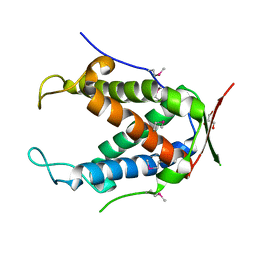 | |
5V0I
 
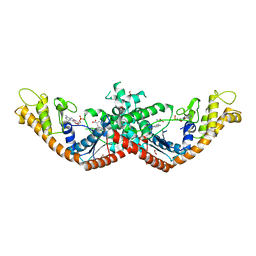 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan | | 分子名称: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, TRYPTOPHAN, ... | | 著者 | Maltseva, N, Kim, Y, Mulligan, R, Grimshaw, S.G, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-28 | | 公開日 | 2017-03-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan
To Be Published
|
|
5TFP
 
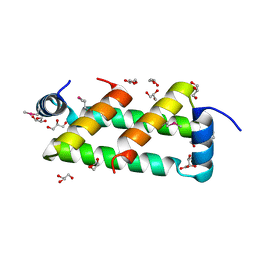 | |
5UZS
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P200 | | 分子名称: | 1,2-ETHANEDIOL, 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-27 | | 公開日 | 2017-03-22 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.367 Å) | | 主引用文献 | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P200
To Be Published
|
|
5UJW
 
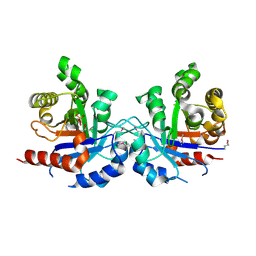 | | Crystal structure of triosephosphate isomerase from Francisella tularensis subsp. tularensis SCHU S4 | | 分子名称: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, CITRIC ACID, ... | | 著者 | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-01-19 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of triosephosphate isomerase from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
5UZE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P182 | | 分子名称: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | 著者 | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-02-26 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P182
To Be Published
|
|
5VSV
 
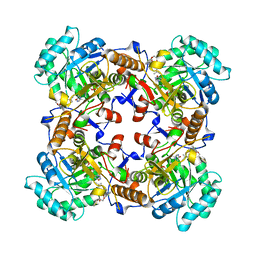 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P225 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, {2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenoxy}acetic acid | | 著者 | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-05-12 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.205 Å) | | 主引用文献 | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P225
To Be Published
|
|
7K0G
 
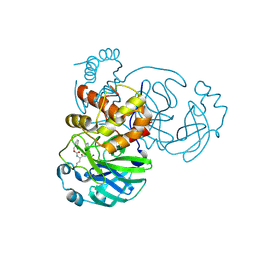 | | 1.85 A resolution structure of SARS-CoV 3CL protease in complex with deuterated GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Replicase polyprotein 1a | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-09-04 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K0E
 
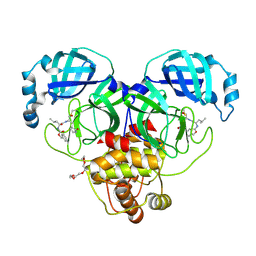 | | 1.90 A resolution structure of SARS-CoV-2 3CL protease in complex with deuterated GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-09-04 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K0H
 
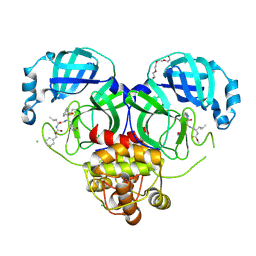 | | 1.70 A resolution structure of SARS-CoV 3CL protease in complex with a deuterated GC376 alpha-ketoamide analog (compound 5) | | 分子名称: | CHLORIDE ION, N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide, Replicase polyprotein 1a, ... | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-09-04 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | 分子名称: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | 著者 | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-07 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
3IGJ
 
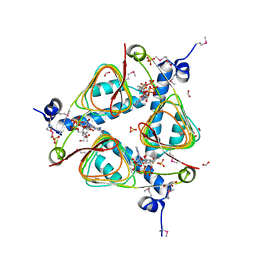 | | Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis | | 分子名称: | ACETYL COENZYME *A, FORMIC ACID, GLYCEROL, ... | | 著者 | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-07-27 | | 公開日 | 2009-08-04 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis
To be Published
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | 分子名称: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | 著者 | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-08-07 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JQE
 
 | | Structure of an extracellular fragment of EsaA from Streptococcus gallolyticus | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, ESAT-6/WXG100 secretion system protein | | 著者 | Klein, T.A, Grebenc, D.W, Kim, Y, Whitney, J.C. | | 登録日 | 2020-08-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2021-02-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of the Extracellular Region of the Bacterial Type VIIb Secretion System Subunit EsaA.
Structure, 29, 2021
|
|
7K0F
 
 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with a deuterated GC376 alpha-ketoamide analog (compound 5) | | 分子名称: | 3C-like proteinase, N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide, TETRAETHYLENE GLYCOL | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-09-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3IQT
 
 | | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073. | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Signal transduction histidine-protein kinase barA | | 著者 | Cuff, M.E, Rakowski, E, Kim, Y, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-08-20 | | 公開日 | 2009-09-22 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073.
TO BE PUBLISHED
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | 分子名称: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | 著者 | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-20 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3HLV
 
 | | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and 16-alpha-hydroxy-estrone ((8S,9R,13S,14R,16R)-3,16-dihydroxy-13-methyl-7,8,9,11,12,14,15, 16-octahydro-6H-cyclopenta[a]phenanthren-17-one | | 分子名称: | (9beta,13alpha,16beta)-3,16-dihydroxyestra-1,3,5(10)-trien-17-one, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Rajan, S.S, Kim, Y, Vanek, K, Joachimiak, A, Greene, G.L. | | 登録日 | 2009-05-28 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of human alphaER-LBD in complex with GRIP peptide and 16alpha-hydroxyestrone
To be Published
|
|
3HM1
 
 | | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15,16-octahydro-6H-cyclopenta[a]phenanthren-17-one) | | 分子名称: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Rajan, S.S, Kim, Y, Vanek, K, Liwanag, M, Joachimiak, A, Greene, G.L. | | 登録日 | 2009-05-28 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15, 16-octahydro-6H-cyclopenta[a]phenanthren-17-one)
To be Published
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | 分子名称: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | 著者 | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LKS
 
 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2f | | 分子名称: | (1R,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | 登録日 | 2021-02-02 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKT
 
 | | 1.50 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2k | | 分子名称: | (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | 著者 | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | 登録日 | 2021-02-02 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
