8EO7
 
 | | Crystal structure of metagenomic beta-lactamase LRA-5 Y69Q/V166E mutant at 2.15 Angstrom resolution | | 分子名称: | beta-lactamase | | 著者 | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | 登録日 | 2022-10-02 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
1FAV
 
 | | THE STRUCTURE OF AN HIV-1 SPECIFIC CELL ENTRY INHIBITOR IN COMPLEX WITH THE HIV-1 GP41 TRIMERIC CORE | | 分子名称: | HIV-1 ENVELOPE PROTEIN CHIMERA, PROTEIN (TRANSMEMBRANE GLYCOPROTEIN) | | 著者 | Zhou, G, Ferrer, M, Chopra, R, Strassmaier, T, Weissenhorn, W, Skehel, J.J, Oprian, D, Schreiber, S.L, Harrison, S.C, Wiley, D.C. | | 登録日 | 2000-07-13 | | 公開日 | 2000-08-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of an HIV-1 specific cell entry inhibitor in complex with the HIV-1 gp41 trimeric core.
Bioorg.Med.Chem., 8, 2000
|
|
1FCA
 
 | |
8EZV
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2022-11-01 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
1F89
 
 | | Crystal structure of Saccharomyces cerevisiae Nit3, a member of branch 10 of the nitrilase superfamily | | 分子名称: | 32.5 KDA PROTEIN YLR351C | | 著者 | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2000-06-29 | | 公開日 | 2001-10-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a putative CN hydrolase from yeast
Proteins, 52, 2003
|
|
8EZZ
 
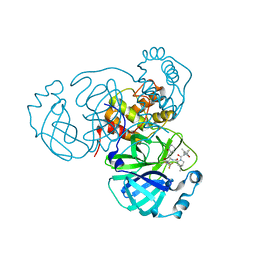 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a2 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-difluoroazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2022-11-01 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8F2D
 
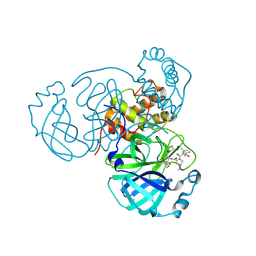 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML4006a | | 分子名称: | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopiperidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2022-11-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
6PK6
 
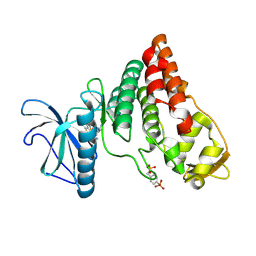 | | Human PRPF4B bound to benzothiophene inhibitor 329 | | 分子名称: | 4-(5-{[(2-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | 著者 | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | 登録日 | 2019-06-28 | | 公開日 | 2019-08-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | to be published
To Be Published
|
|
8EO6
 
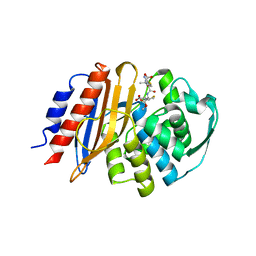 | | Crystal structure of metagenomic class A beta-lactamase precursor LRA-5 in complex with ceftazidime at 2.35 Angstrom resolution | | 分子名称: | ACYLATED CEFTAZIDIME, LRA-5 | | 著者 | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | 登録日 | 2022-10-02 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
8F02
 
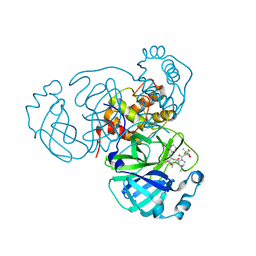 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a4 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-dimethylazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2022-11-01 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
1F8F
 
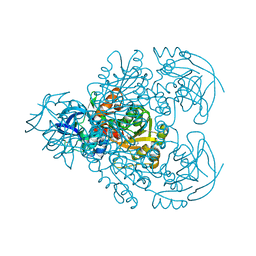 | | CRYSTAL STRUCTURE OF BENZYL ALCOHOL DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS | | 分子名称: | BENZYL ALCOHOL DEHYDROGENASE, ETHANOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Beauchamp, J.C, Gillooly, D, Warwicker, J, Fewson, C.A, Lapthorn, A.J. | | 登録日 | 2000-06-30 | | 公開日 | 2003-07-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Benzyl Alcohol Dehydrogenase from Acinetobacter calcoaceticus
To be Published
|
|
8F2C
 
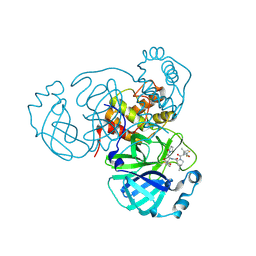 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML3006a | | 分子名称: | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopyrrolidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Westberg, M, Fernandez, D, Lin, M.Z. | | 登録日 | 2022-11-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
1F97
 
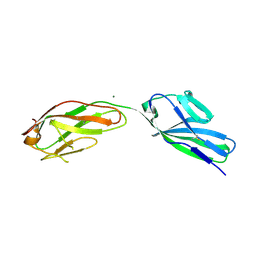 | | SOLUBLE PART OF THE JUNCTION ADHESION MOLECULE FROM MOUSE | | 分子名称: | JUNCTION ADHESION MOLECULE, MAGNESIUM ION | | 著者 | Kostrewa, D, Brockhaus, M, D'Arcy, A, Dale, G, Bazzoni, G, Dejana, E, Winkler, F, Hennig, M. | | 登録日 | 2000-07-07 | | 公開日 | 2001-01-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif.
EMBO J., 20, 2001
|
|
1FQ1
 
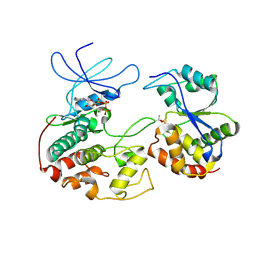 | | CRYSTAL STRUCTURE OF KINASE ASSOCIATED PHOSPHATASE (KAP) IN COMPLEX WITH PHOSPHO-CDK2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-DEPENDENT KINASE INHIBITOR 3, ... | | 著者 | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | 登録日 | 2000-09-01 | | 公開日 | 2001-05-09 | | 最終更新日 | 2018-03-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
4XX5
 
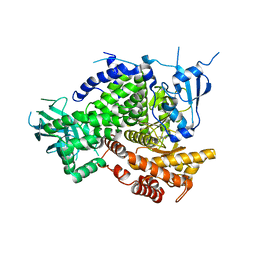 | | Structure of PI3K gamma in complex with an inhibitor | | 分子名称: | N-[5-(5-methoxypyridin-3-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | 著者 | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | 登録日 | 2015-01-29 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
1FBY
 
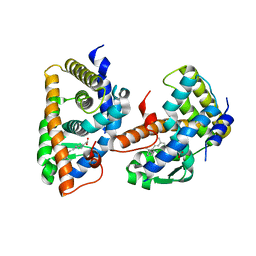 | | CRYSTAL STRUCTURE OF THE HUMAN RXR ALPHA LIGAND BINDING DOMAIN BOUND TO 9-CIS RETINOIC ACID | | 分子名称: | (9cis)-retinoic acid, RETINOIC ACID RECEPTOR RXR-ALPHA | | 著者 | Egea, P.F, Mitschler, A, Rochel, N, Ruff, M, Chambon, P, Moras, D. | | 登録日 | 2000-07-17 | | 公開日 | 2000-07-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of the human RXRalpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid.
EMBO J., 19, 2000
|
|
1FBM
 
 | | ASSEMBLY DOMAIN OF CARTILAGE OLIGOMERIC MATRIX PROTEIN IN COMPLEX WITH ALL-TRANS RETINOL | | 分子名称: | PROTEIN (CARTILAGE OLIGOMERIC MATRIX PROTEIN), RETINOL | | 著者 | Guo, Y, Bozic, D, Malashkevich, V.N, Kammerer, R.A, Schulthess, T. | | 登録日 | 2000-07-16 | | 公開日 | 2000-08-02 | | 最終更新日 | 2018-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | All-trans retinol, vitamin D and other hydrophobic compounds bind in the axial pore of the five-stranded coiled-coil domain of cartilage oligomeric matrix protein.
EMBO J., 17, 1998
|
|
6ZRB
 
 | |
1FBZ
 
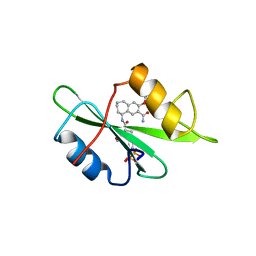 | | Structure-based design of a novel, osteoclast-selective, nonpeptide Src SH2 inhibitor with in vivo anti-resorptive activity | | 分子名称: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK, {4-[2-ACETYLAMINO-2-(3-CARBAMOYL-2-CYCLOHEXYLMETHOXY-6,7,8,9-TETRAHYDRO-5H-BENZOCYCLOHEPTEN-5YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENYL}-PHOSPHONIC ACID | | 著者 | Shakespeare, W, Yang, M, Bohacek, R, Cerasoli, F, Stebbis, K, Sundaramoorthi, R, Vu, C, Pradeepan, S, Metcalf, C, Haraldson, C, Merry, T, Dalgarno, D, Narula, S, Hatada, M, Lu, X, Van Schravendijk, M.R, Adams, S, Violette, S, Smith, J, Guan, W, Bartlett, C, Herson, J, Iuliucci, J, Weigele, M, Sawyer, T. | | 登録日 | 2000-07-17 | | 公開日 | 2000-08-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-based design of an osteoclast-selective, nonpeptide src homology 2 inhibitor with in vivo antiresorptive activity.
Proc.Natl.Acad.Sci.Usa, 97, 2000
|
|
6P24
 
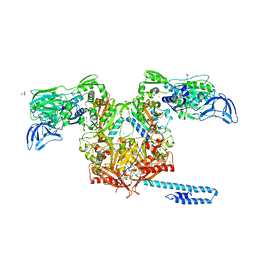 | | Escherichia coli tRNA synthetase | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Kahne, D, Baidin, V, Owens, T.W. | | 登録日 | 2019-05-20 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Simple Secondary Amines Inhibit Growth of Gram-Negative Bacteria through Highly Selective Binding to Phenylalanyl-tRNA Synthetase.
J.Am.Chem.Soc., 143, 2021
|
|
6P44
 
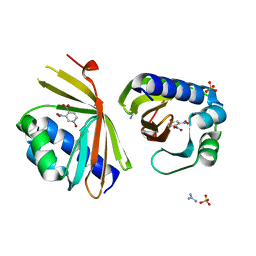 | | Crystal Structure of Ketosteroid Isomerase D38N mutant from Mycobacterium hassiacum (mhKSI) bound to 3,4-dinitrophenol | | 分子名称: | 3,4-dinitrophenol, GUANIDINE, SULFATE ION, ... | | 著者 | Yabukarski, F, Doukov, T, Pinney, M, Herschlag, D. | | 登録日 | 2019-05-25 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.251 Å) | | 主引用文献 | Parallel molecular mechanisms for enzyme temperature adaptation.
Science, 371, 2021
|
|
8F7H
 
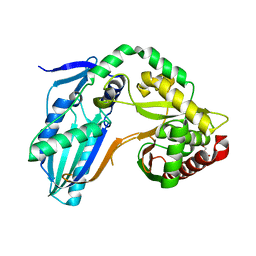 | | The condensation domain of surfactin A synthetase C variant 18b in space group P212121 | | 分子名称: | GLYCEROL, Surfactin synthetase | | 著者 | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | 登録日 | 2022-11-18 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
4XUG
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with 2-({[4-(Trifluoromethoxy)Phenyl]Sulfonyl}Amino)Ethyl Dihydrogen Phosphate (F9F) inhibitor in the alpha site and ammonium ion in the metal coordination site. | | 分子名称: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, AMMONIUM ION, ... | | 著者 | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | 登録日 | 2015-01-25 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with 2-({[4-(Trifluoromethoxy)Phenyl]Sulfonyl}Amino)Ethyl Dihydrogen Phosphate (F9F) inhibitor in the alpha site and ammonium ion in the metal coordination site.
To Be Published
|
|
8F7I
 
 | | The condensation domain of surfactin A synthetase C variant 18b in space group P43212 | | 分子名称: | GLYCEROL, Surfactin synthetase | | 著者 | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, M. | | 登録日 | 2022-11-18 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
8F7G
 
 | | The condensation domain of surfactin A synthetase C in space group P212121 | | 分子名称: | GLYCEROL, Surfactin synthetase | | 著者 | Frota, N.F, Pistofidis, A, Folger, I.B, Hilvert, D, Schmeing, T.M. | | 登録日 | 2022-11-18 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | High-throughput reprogramming of an NRPS condensation domain.
Nat.Chem.Biol., 20, 2024
|
|
