6JWG
 
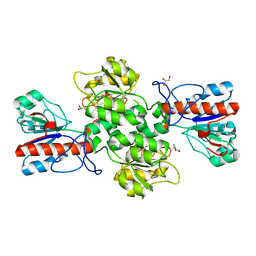 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Formate dehydrogenase, GLYCEROL | | 著者 | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | 登録日 | 2019-04-20 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.081 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
5CCP
 
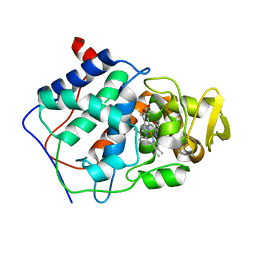 | | HISTIDINE 52 IS A CRITICAL RESIDUE FOR RAPID FORMATION OF CYTOCHROME C PEROXIDASE COMPOUND I | | 分子名称: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Brown, K, Shaw, A, Miller, M.A, Kraut, J. | | 登録日 | 1993-06-07 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Histidine 52 is a critical residue for rapid formation of cytochrome c peroxidase compound I.
Biochemistry, 32, 1993
|
|
3ME9
 
 | | Crystal structure of SGF29 in complex with H3K4me3 peptide | | 分子名称: | GLYCEROL, Histone H3, SAGA-associated factor 29 homolog, ... | | 著者 | Bian, C, Tempel, W, Xu, C, Guo, Y, Dong, A, Crombet, L, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-03-31 | | 公開日 | 2010-04-28 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
3TZD
 
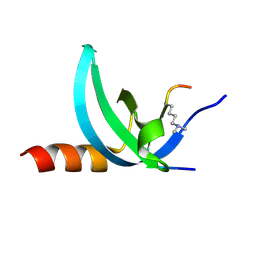 | | Crystal structure of the complex of Human Chromobox Homolog 3 (CBX3) | | 分子名称: | Chromobox protein homolog 3, Histone H1.4 | | 著者 | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | 登録日 | 2011-09-27 | | 公開日 | 2012-03-07 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
3SRQ
 
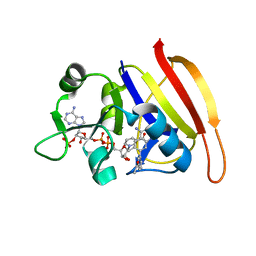 | |
3MEA
 
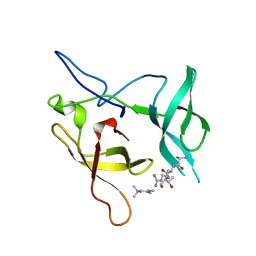 | | Crystal structure of the SGF29 in complex with H3K4me3 | | 分子名称: | Histone H3, SAGA-associated factor 29 homolog | | 著者 | Bian, C, Xu, C, Tempel, W, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-03-31 | | 公開日 | 2010-04-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
5DD6
 
 | |
3SRS
 
 | |
5V0Y
 
 | |
3SRU
 
 | |
4RKG
 
 | |
5DD3
 
 | |
7LFM
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, TRICLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | 著者 | Tomchick, D.R, Deisenhofer, J, Shen, S. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFL
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, VAL MUTANT, MONOCLINIC CELL, REFINED AT 1.60 ANGSTROMS RESOLUTION | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | 著者 | Tomchick, D.R, Deisenhofer, J, Shen, S. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2022-12-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFK
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, THR MUTANT, REFINED AT 1.60 ANGSTROMS RESOLUTION | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | 著者 | Tomchick, D.R, Deisenhofer, J, Shen, S. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFJ
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE, ALA MUTANT, REFINED AT 1.70 ANGSTROMS RESOLUTION | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | 著者 | Tomchick, D.R, Deisenhofer, J, Shen, S. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
7LFI
 
 | | MODEL OF MHC CLASS Ib H2-M3 WITH MOUSE ND1 N-TERMINAL HEPTAPEPTIDE REFINED AT 1.70 ANGSTROMS RESOLUTION | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Heptapeptide from NADH-ubiquinone oxidoreductase chain 1, ... | | 著者 | Tomchick, D.R, Deisenhofer, J, Shen, S. | | 登録日 | 2021-01-17 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
J.Biomol.Struct.Dyn., 40, 2022
|
|
3EMG
 
 | |
4JMH
 
 | |
5YZW
 
 | | Crystal structure of p204 HINb domain | | 分子名称: | Ifi204 | | 著者 | Jin, T. | | 登録日 | 2017-12-15 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
4N39
 
 | |
4Q1B
 
 | |
4Q1D
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the inhibitor 9 {2-{[(1R)-1-{2-[3-(2-fluoroethoxy)-4-methoxyphenyl]-5-propyl-1,3-thiazol-4-yl}ethyl]sulfanyl}pyrimidine-4,6-diamine} | | 分子名称: | (R)-2-((1-(2-(3-(2-fluoroethoxy)-4-methoxyphenyl)-5-propylthiazol-4-yl)ethyl)thio)pyrimidine-4,6-diamine, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | 著者 | Nomme, J, Lavie, A. | | 登録日 | 2014-04-03 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-guided development of deoxycytidine kinase inhibitors with nanomolar affinity and improved metabolic stability.
J.Med.Chem., 57, 2014
|
|
4N3B
 
 | | Crystal Structure of human O-GlcNAc Transferase bound to a peptide from HCF-1 pro-repeat2(1-26)E10Q and UDP-5SGlcNAc | | 分子名称: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Host cell factor 1, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | 著者 | Lazarus, M.B, Herr, W, Walker, S. | | 登録日 | 2013-10-06 | | 公開日 | 2014-01-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | HCF-1 is cleaved in the active site of O-GlcNAc transferase.
Science, 342, 2013
|
|
4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | 分子名称: | CITRIC ACID, DNA (167-mer), H5, ... | | 著者 | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | 登録日 | 2014-06-11 | | 公開日 | 2015-07-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.503 Å) | | 主引用文献 | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
