7F8J
 
 | | Cryo-EM structure of human pannexin-1 in a nanodisc | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | 著者 | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | 登録日 | 2021-07-02 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8N
 
 | | Human pannexin-1 showing a conformational change in the N-terminal domain and blocked pore | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | 著者 | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | 登録日 | 2021-07-02 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | 著者 | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | 登録日 | 2021-07-02 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
2D1V
 
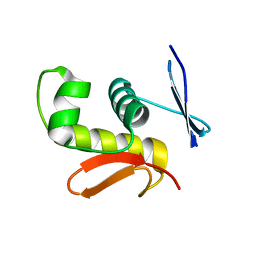 | | Crystal structure of DNA-binding domain of Bacillus subtilis YycF | | 分子名称: | Transcriptional regulatory protein yycF | | 著者 | Okajima, T, Okada, A, Watanabe, T, Yamamoto, K, Tanizawa, K, Utsumi, R. | | 登録日 | 2005-09-01 | | 公開日 | 2006-09-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Response regulator YycF essential for bacterial growth: X-ray crystal structure of the DNA-binding domain and its PhoB-like DNA recognition motif
Febs Lett., 582, 2008
|
|
3A8Q
 
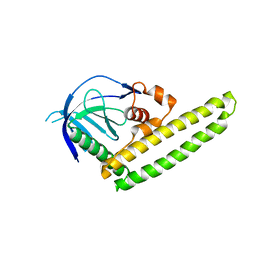 | | Low-resolution crystal structure of the Tiam2 PHCCEx domain | | 分子名称: | T-lymphoma invasion and metastasis-inducing protein 2 | | 著者 | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | 登録日 | 2009-10-07 | | 公開日 | 2009-11-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
4XRE
 
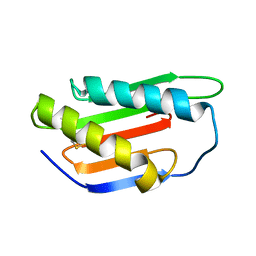 | | Crystal structure of Gnk2 complexed with mannose | | 分子名称: | Antifungal protein ginkbilobin-2, alpha-D-mannopyranose | | 著者 | Miyakawa, T, Hatano, K, Miyauchi, Y, Suwa, Y, Sawano, Y, Tanokura, M. | | 登録日 | 2015-01-21 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.597 Å) | | 主引用文献 | A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity.
Plant Physiol., 166, 2014
|
|
4Y6I
 
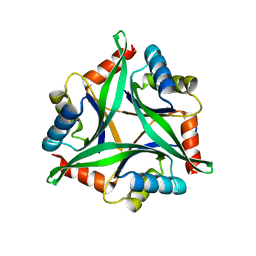 | |
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | 分子名称: | T-lymphoma invasion and metastasis-inducing protein 2 | | 著者 | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | 登録日 | 2009-10-07 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
4Y65
 
 | |
3A8N
 
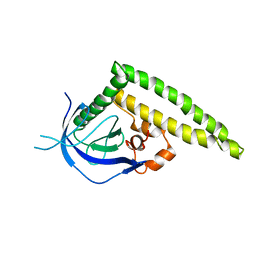 | | Crystal structure of the Tiam1 PHCCEx domain | | 分子名称: | T-lymphoma invasion and metastasis-inducing protein 1 | | 著者 | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | 登録日 | 2009-10-07 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3AA9
 
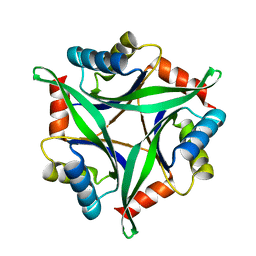 | | Crystal Structure Analysis of the Mutant CutA1 (E61V) from E. coli | | 分子名称: | Divalent-cation tolerance protein cutA | | 著者 | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | 登録日 | 2009-11-12 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
3IXZ
 
 | | Pig gastric H+/K+-ATPase complexed with aluminium fluoride | | 分子名称: | Potassium-transporting ATPase alpha, Potassium-transporting ATPase subunit beta | | 著者 | Abe, K, Tani, K, Nishizawa, T, Fujiyoshi, Y. | | 登録日 | 2009-03-09 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (6.5 Å) | | 主引用文献 | Inter-subunit interaction of gastric H+,K+-ATPase prevents reverse reaction of the transport cycle
Embo J., 28, 2009
|
|
3AA8
 
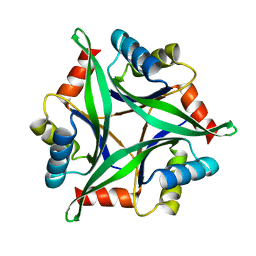 | | Crystal Structure Analysis of the Mutant CutA1 (S11V/E61V) from E. coli | | 分子名称: | Divalent-cation tolerance protein cutA | | 著者 | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | 登録日 | 2009-11-12 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
8YHR
 
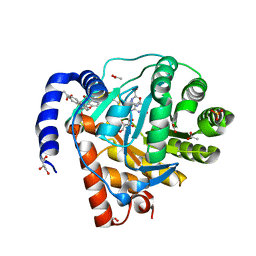 | | DHODH in complex with furocoumavirin | | 分子名称: | 4-methyl-8-[(S)-oxidanyl(phenyl)methyl]-9-phenyl-furo[2,3-h]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | 著者 | Hara, K, Okumura, H, Nakahara, M, Sato, M, Hashimoto, H, Osada, H, Watanabe, K. | | 登録日 | 2024-02-28 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Functional Analyses of Inhibition of Human Dihydroorotate Dehydrogenase by Antiviral Furocoumavirin.
Biochemistry, 63, 2024
|
|
8AU4
 
 | | Structural insights reveal a heterotetramer between oncogenic K-Ras4BG12V and Rgl2, a RalA/B activator | | 分子名称: | Ral guanine nucleotide dissociation stimulator-like 2 | | 著者 | Tariq, M, Ikeya, T, Togashi, N, Fairall, L, Alejo, C.B, Kamei, S, Alonso, B.R, Campillo, M.A.M, Hudson, A, Ito, Y, Schwabe, J, Dominguez, C, Tanaka, K. | | 登録日 | 2022-08-25 | | 公開日 | 2023-08-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
8B69
 
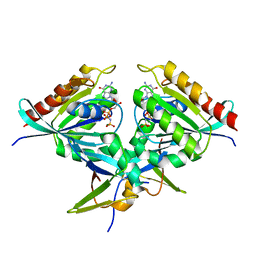 | | Heterotetramer of K-Ras4B(G12V) and Rgl2(RBD) | | 分子名称: | Isoform 2B of GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | 著者 | Tariq, M, Fairall, L, Romartinez-Alonso, B, Dominguez, C, Schwabe, J.W.R, Tanaka, K. | | 登録日 | 2022-09-26 | | 公開日 | 2023-08-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
3AH6
 
 | | Remarkable improvement of the heat stability of CutA1 from E.coli by rational protein designing | | 分子名称: | Divalent-cation tolerance protein cutA | | 著者 | Matsuura, Y, Tanaka, T, Bagautdinov, B, Kunishima, N, Yutani, K. | | 登録日 | 2010-04-15 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Remarkable improvement in the heat stability of CutA1 from Escherichia coli by rational protein design
J.Biochem., 148, 2010
|
|
6IZP
 
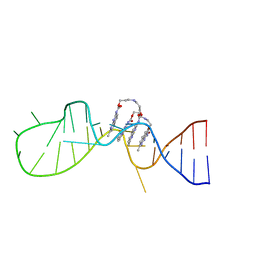 | | Solution structure of the complex of naphthyridine carbamate dimer and an RNA with UGGAA-UGGAA pentad | | 分子名称: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, RNA (29-MER) | | 著者 | Nagano, K, Shibata, T, Nakatani, K, Kawai, G. | | 登録日 | 2018-12-20 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun, 12, 2021
|
|
1WQO
 
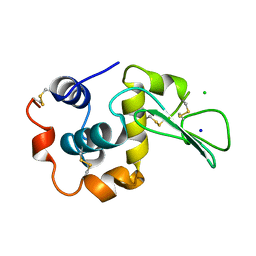 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQM
 
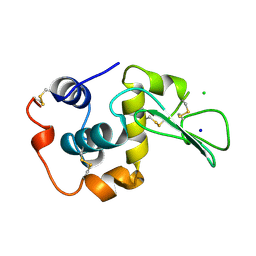 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQP
 
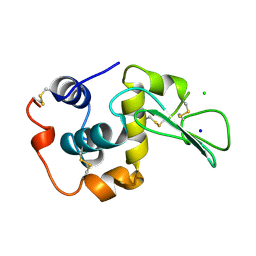 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQR
 
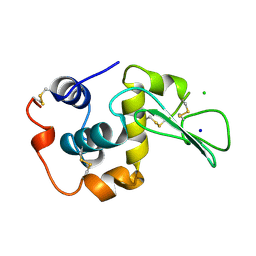 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQN
 
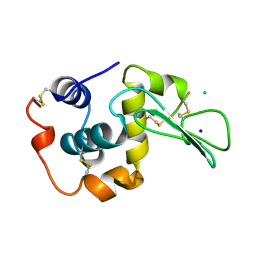 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
1WQQ
 
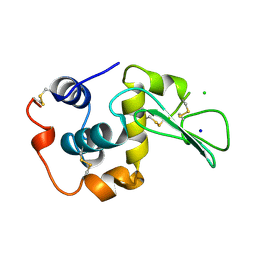 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME | | 分子名称: | CHLORIDE ION, LYSOZYME, SODIUM ION | | 著者 | Yamagata, Y, Kubota, M, Sumikawa, Y, Funahashi, J, Fujii, S, Yutani, K. | | 登録日 | 1998-02-09 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six tyrosine --> phenylalanine mutants.
Biochemistry, 37, 1998
|
|
2ZWB
 
 | | Neutron crystal structure of wild type human lysozyme in D2O | | 分子名称: | Lysozyme C | | 著者 | Chiba-Kamoshida, K, Matsui, T, Chatake, T, Ohhara, T, Ostermann, A, Tanaka, I, Yutani, K, Niimura, N. | | 登録日 | 2008-12-02 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | NEUTRON DIFFRACTION (1.8 Å) | | 主引用文献 | Site-specific softening of peptide bonds by localized deuterium observed by neutron crystallography of human lysozyme
To be Published
|
|
