7YHW
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.12.1 RBD in complex with human ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-07-14 | | 公開日 | 2023-07-19 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7DSY
 
 | | Crystal Structure of RNase L in complex with KM05073 | | 分子名称: | 1-chloranyl-3-methylsulfinyl-6,7-dihydro-5H-2-benzothiophen-4-one, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, ... | | 著者 | Tang, J, Huang, H. | | 登録日 | 2021-01-03 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.651 Å) | | 主引用文献 | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7OLB
 
 | | Crystal structure of Pab-AGOG, an 8-oxoguanine DNA glycosylase from Pyrococcus abyssi | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | 登録日 | 2021-05-19 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OLI
 
 | | Crystal structure of Pab-AGOG in complex with 8-oxoguanosine | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXY-8-OXOGUANOSINE, N-glycosylase/DNA lyase | | 著者 | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | 登録日 | 2021-05-20 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OUE
 
 | | Crystal structure of a trapped Pab-AGOG/single-standed DNA covalent intermediate | | 分子名称: | DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), N-glycosylase/DNA lyase, PHOSPHATE ION, ... | | 著者 | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | 登録日 | 2021-06-11 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OU3
 
 | | Crystal structure of Tga-AGOG, an 8-oxoguanine DNA glycosylase from Thermococcus gammatolerans | | 分子名称: | CHLORIDE ION, GLYCEROL, N-glycosylase/DNA lyase | | 著者 | Coste, F, Confalonieri, F, Castaing, B. | | 登録日 | 2021-06-11 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
6OYL
 
 | | The structure of the PP2A B56 subunit KIF4A complex | | 分子名称: | Chromosome-associated kinesin KIF4A, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | 著者 | Wang, X, Page, R, Peti, W. | | 登録日 | 2019-05-14 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | A dynamic charge-charge interaction modulates PP2A:B56 substrate recruitment.
Elife, 9, 2020
|
|
7OY7
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing cytosine opposite to lesion) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | 著者 | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | 登録日 | 2021-06-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7P0W
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing thymine opposite to lesion) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | 著者 | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | 登録日 | 2021-06-30 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
5DDW
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | 分子名称: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Xu, J, Feng, Z, Liu, J. | | 登録日 | 2015-08-25 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
7DTS
 
 | | Crystal structure of RNase L in complex with AC40357 | | 分子名称: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, Ribonuclease L, ... | | 著者 | Tang, J, Huang, H. | | 登録日 | 2021-01-06 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7ELW
 
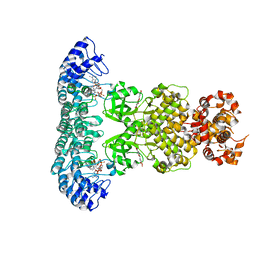 | | Crystal structure of RNase L in complex with Myricetin | | 分子名称: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, PHOSPHATE ION, Ribonuclease L, ... | | 著者 | Tang, J, Huang, H. | | 登録日 | 2021-04-12 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
6LP9
 
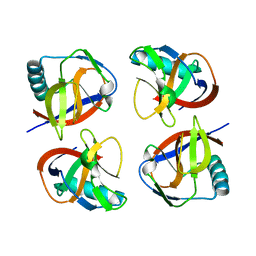 | | the protein of cat virus | | 分子名称: | nsp1 protein | | 著者 | Shen, Z, Peng, G.Q. | | 登録日 | 2020-01-09 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Structural and Biological Basis of Alphacoronavirus nsp1 Associated with Host Proliferation and Immune Evasion.
Viruses, 12, 2020
|
|
8X85
 
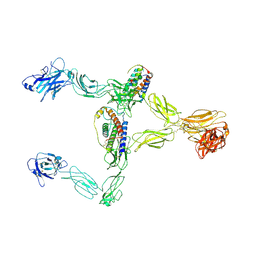 | | Structure of leptin-LepR dimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, Leptin receptor, ... | | 著者 | Xie, Y.F, Shang, G.J, Qi, J.X, Gao, G.F. | | 登録日 | 2023-11-27 | | 公開日 | 2024-08-07 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural plasticity of human leptin binding to its receptor LepR
Hlife, 1, 2023
|
|
6R0K
 
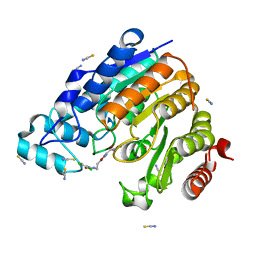 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with a inhibitor SS208 | | 分子名称: | 5-[2-[(3,4-dichlorophenyl)carbonylamino]ethyl]-~{N}-oxidanyl-1,2-oxazole-3-carboxamide, Histone deacetylase 6, POTASSIUM ION, ... | | 著者 | Barinka, C, Ustinova, K, Motlova, L, Pavlicek, J. | | 登録日 | 2019-03-13 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Discovery of a New Isoxazole-3-hydroxamate-Based Histone Deacetylase 6 Inhibitor SS-208 with Antitumor Activity in Syngeneic Melanoma Mouse Models.
J.Med.Chem., 62, 2019
|
|
2XK9
 
 | | Structural analysis of checkpoint kinase 2 (Chk2) in complex with inhibitor PV1533 | | 分子名称: | CHECKPOINT KINASE 2, N-{4-[(1E)-N-(N-hydroxycarbamimidoyl)ethanehydrazonoyl]phenyl}-7-nitro-1H-indole-2-carboxamide | | 著者 | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Shoemaker, R.H, Pommier, Y, Waugh, D.S. | | 登録日 | 2010-07-07 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
8GRY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with rat ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | 登録日 | 2022-09-03 | | 公開日 | 2023-07-19 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8H06
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with human ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-09-28 | | 公開日 | 2023-07-19 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-10-12 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
4YFF
 
 | | TNNI3K complexed with inhibitor 2 | | 分子名称: | 3-[(5-bromo-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-N-methyl-4-(morpholin-4-yl)benzenesulfonamide, Serine/threonine-protein kinase TNNI3K | | 著者 | Shewchuk, L.M, Wang, L, Lawhorn, B.G. | | 登録日 | 2015-02-25 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Identification of Purines and 7-Deazapurines as Potent and Selective Type I Inhibitors of Troponin I-Interacting Kinase (TNNI3K).
J.Med.Chem., 58, 2015
|
|
6V5A
 
 | |
4YFI
 
 | | TNNI3K complexed with inhibitor 1 | | 分子名称: | N-methyl-3-(9H-purin-6-ylamino)benzenesulfonamide, Serine/threonine-protein kinase TNNI3K | | 著者 | Shewchuk, L.M, Wang, L, Lawhorn, B.G. | | 登録日 | 2015-02-25 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Identification of Purines and 7-Deazapurines as Potent and Selective Type I Inhibitors of Troponin I-Interacting Kinase (TNNI3K).
J.Med.Chem., 58, 2015
|
|
6LVV
 
 | | N, N-dimethylformamidase | | 分子名称: | 1,2-ETHANEDIOL, FE (III) ION, N,N-dimethylformamidase large subunit, ... | | 著者 | Arya, C.K, Ramaswamy, S, Kutti, R.V, Gurunath, R. | | 登録日 | 2020-02-05 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4YZ9
 
 | |
4YZC
 
 | |
