3RC0
 
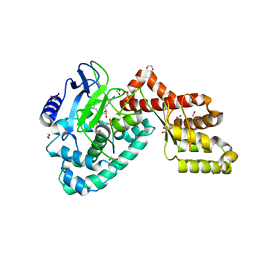 | | Human SETD6 in complex with RelA Lys310 peptide | | 分子名称: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | 著者 | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | 登録日 | 2011-03-30 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
3QXY
 
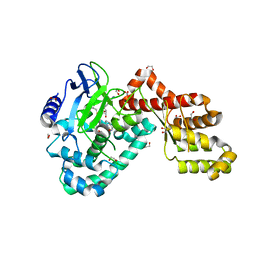 | | Human SETD6 in complex with RelA Lys310 | | 分子名称: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | 著者 | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | 登録日 | 2011-03-02 | | 公開日 | 2011-05-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
7QCQ
 
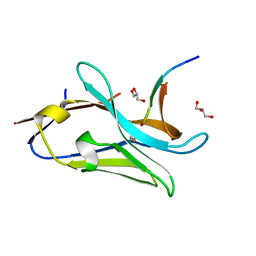 | | VHH Z70 in interaction with PHF6 Tau peptide | | 分子名称: | GLYCEROL, Tau 301-312, VHH Z70 | | 著者 | Danis, C, Dupre, E, Zejneli, O, Caillierez, R, Arrial, A, Begard, S, Loyens, A, Mortelecque, J, Cantrelle, F.-X, Hanoulle, X, Rain, J.-C, Colin, M, Buee, L, Landrieu, I. | | 登録日 | 2021-11-25 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Inhibition of Tau seeding by targeting Tau nucleation core within neurons with a single domain antibody fragment.
Mol.Ther., 30, 2022
|
|
3SWC
 
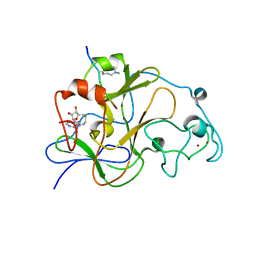 | | GLP (G9a-like protein) SET domain in complex with Dnmt3aK44me2 peptide | | 分子名称: | DNA (cytosine-5)-methyltransferase 3A, Histone-lysine N-methyltransferase EHMT1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Chang, Y, Horton, J.R, Zhang, X, Cheng, X. | | 登録日 | 2011-07-13 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.332 Å) | | 主引用文献 | MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a.
Nat Commun, 2, 2011
|
|
5CB3
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5COA
 
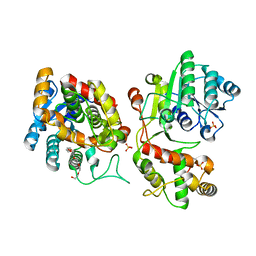 | | Crystal structure of iridoid synthase at 2.2-angstrom resolution | | 分子名称: | HEXAETHYLENE GLYCOL, Iridoid synthase, SULFATE ION | | 著者 | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
5COB
 
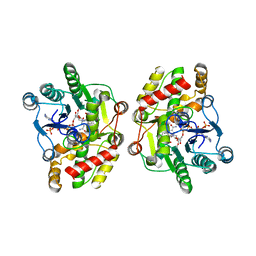 | | Crystal structure of iridoid synthase in complex with NADP+ and 8-oxogeranial at 2.65-angstrom resolution | | 分子名称: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5CMS
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, SULFATE ION | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-07-17 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
7RFO
 
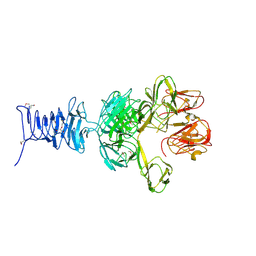 | | SeMet Tailspike protein 4 (TSP4) phage CBA120, residues 1-335, obtained in the presence of LiSO4 | | 分子名称: | Tailspike protein | | 著者 | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | 登録日 | 2021-07-14 | | 公開日 | 2022-02-23 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7REJ
 
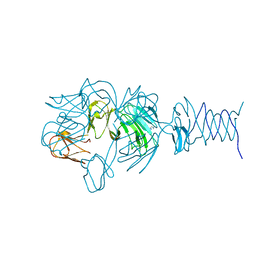 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-335, obtained in the presence of NaK-Tartrate | | 分子名称: | IMIDAZOLE, Tailspike protein | | 著者 | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | 登録日 | 2021-07-13 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7RFV
 
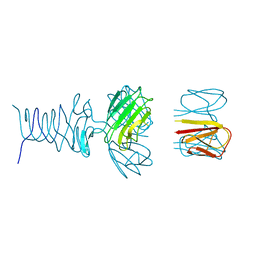 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-250, obtained in the presence of PEG8000 | | 分子名称: | Tailspike protein | | 著者 | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | 登録日 | 2021-07-14 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
3QO2
 
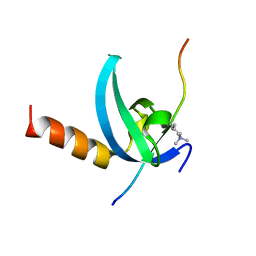 | | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9 | | 分子名称: | 1,2-ETHANEDIOL, Histone H3 peptide, M-phase phosphoprotein 8 | | 著者 | Chang, Y, Horton, J.R, Bedford, M.T, Zhang, X, Cheng, X. | | 登録日 | 2011-02-09 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding.
J.Mol.Biol., 408, 2011
|
|
7UDB
 
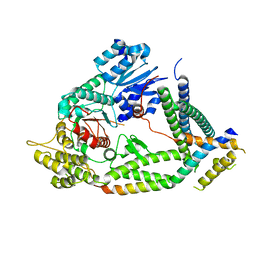 | | Cryo-EM structure of a synaptobrevin-Munc18-1-syntaxin-1 complex class 2 | | 分子名称: | Synaptobrevin-2, Syntaxin-1A, Syntaxin-binding protein 1 | | 著者 | Rizo, J, Bai, X, Stepien, K.P, Xu, J, Zhang, X. | | 登録日 | 2022-03-18 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex.
Sci Adv, 8, 2022
|
|
7V3P
 
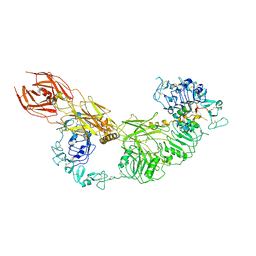 | | Cryo-EM structure of the IGF1R/insulin complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin A chain, Insulin B chain, ... | | 著者 | Zhang, J, Liu, C, Zhang, X, Wei, T, Wu, C. | | 登録日 | 2021-08-11 | | 公開日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of the IGF1R/insulin complex
To Be Published
|
|
7UDC
 
 | | cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex class1 | | 分子名称: | Synaptobrevin-2, Syntaxin-1A, Syntaxin-binding protein 1 | | 著者 | Rizo, J, Bai, X, Stepien, K.P, Xu, J, Zhang, X. | | 登録日 | 2022-03-18 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex.
Sci Adv, 8, 2022
|
|
3SW9
 
 | | GLP (G9a-like protein) SET domain in complex with Dnmt3aK44me0 peptide | | 分子名称: | DNA (cytosine-5)-methyltransferase 3A, Histone-lysine N-methyltransferase EHMT1, SINEFUNGIN, ... | | 著者 | Chang, Y, Horton, J.R, Zhang, X, Cheng, X. | | 登録日 | 2011-07-13 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a.
Nat Commun, 2, 2011
|
|
1DP4
 
 | | DIMERIZED HORMONE BINDING DOMAIN OF THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATRIAL NATRIURETIC PEPTIDE RECEPTOR A, ... | | 著者 | van den Akker, F, Zhang, X, Miyagi, M, Huo, X, Misono, K.S, Yee, V.C. | | 登録日 | 1999-12-23 | | 公開日 | 2000-07-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the dimerized hormone-binding domain of a guanylyl-cyclase-coupled receptor.
Nature, 406, 2000
|
|
6LS0
 
 | |
6LS3
 
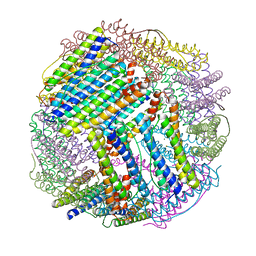 | |
1P9E
 
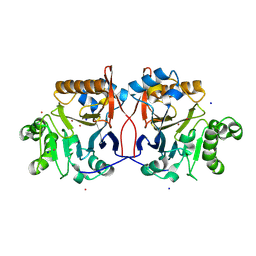 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | 分子名称: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | 著者 | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | 登録日 | 2003-05-11 | | 公開日 | 2004-05-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3
To be Published
|
|
4QNR
 
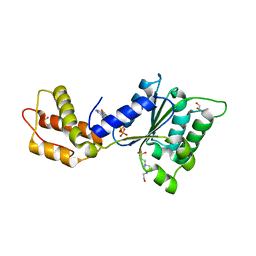 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ATP | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | 著者 | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | 登録日 | 2014-06-18 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.539 Å) | | 主引用文献 | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QOS
 
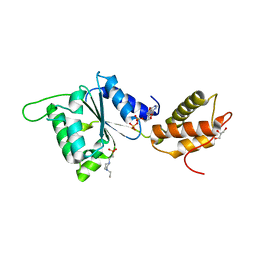 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ADP | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | 登録日 | 2014-06-20 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QNM
 
 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | 著者 | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | 登録日 | 2014-06-18 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.628 Å) | | 主引用文献 | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
6N9Y
 
 | | Atomic structure of Non-Structural protein 1 of bluetongue virus | | 分子名称: | Non-structural protein 1 | | 著者 | Kerviel, A, Ge, P, Lai, M, Jih, J, Boyce, M, Zhang, X, Zhou, Z.H, Roy, P. | | 登録日 | 2018-12-04 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Atomic structure of the translation regulatory protein NS1 of bluetongue virus.
Nat Microbiol, 4, 2019
|
|
