3N9Q
 
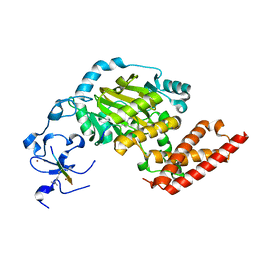 | | ceKDM7A from C.elegans, complex with H3K4me3 peptide, H3K27me2 peptide and NOG | | 分子名称: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | 著者 | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | 登録日 | 2010-05-31 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9N
 
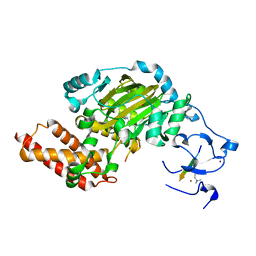 | | ceKDM7A from C.elegans, complex with H3K4me3K9me2 peptide and NOG | | 分子名称: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | 著者 | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | 登録日 | 2010-05-31 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
3N9L
 
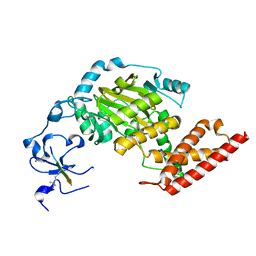 | | ceKDM7A from C.elegans, complex with H3K4me3 peptide and NOG | | 分子名称: | FE (II) ION, Histone H3 peptide, N-OXALYLGLYCINE, ... | | 著者 | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | 登録日 | 2010-05-31 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.796 Å) | | 主引用文献 | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
9U9C
 
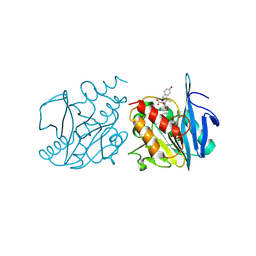 | | Crystal structure of NDM-1 in complex with hydrolyzed amoxicillin | | 分子名称: | (2~{R},4~{S})-2-[(1~{R})-1-[[(2~{R})-2-azanyl-2-(4-hydroxyphenyl)ethanoyl]amino]-2-oxidanyl-2-oxidanylidene-ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | 著者 | Shi, X, Zhang, Q, Liu, W. | | 登録日 | 2025-03-27 | | 公開日 | 2025-07-09 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal structure reveals the hydrophilic R1 group impairs NDM-1-ligand binding via water penetration at L3
J Struct Biol X, 2025
|
|
9N93
 
 | |
9N95
 
 | |
2YQ7
 
 | | Structure of Bcl-xL bound to BimLOCK | | 分子名称: | BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11, GLYCEROL | | 著者 | Smith, B.J, Czabotar, P.E. | | 登録日 | 2012-11-06 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
5BU3
 
 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | 分子名称: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | 著者 | Pan, L, Guo, Y, Liu, J. | | 登録日 | 2015-06-03 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.897 Å) | | 主引用文献 | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
4P6X
 
 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | 分子名称: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | 著者 | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | 登録日 | 2014-03-25 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
3PUQ
 
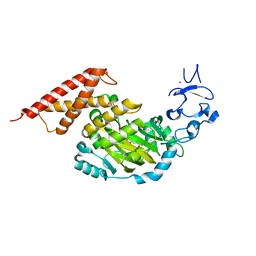 | | CEKDM7A from C.Elegans, complex with alpha-KG | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, GLYCEROL, ... | | 著者 | Yang, Y, Wang, P, Xu, W, Xu, Y. | | 登録日 | 2010-12-06 | | 公開日 | 2011-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
3PUR
 
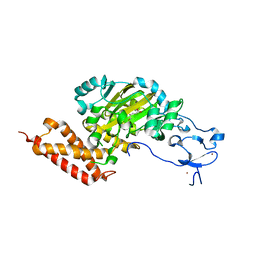 | | CEKDM7A from C.Elegans, complex with D-2-HG | | 分子名称: | (2R)-2-hydroxypentanedioic acid, FE (II) ION, Lysine-specific demethylase 7 homolog, ... | | 著者 | Yang, Y, Wang, P, Xu, W, Xu, Y. | | 登録日 | 2010-12-06 | | 公開日 | 2011-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
5BTU
 
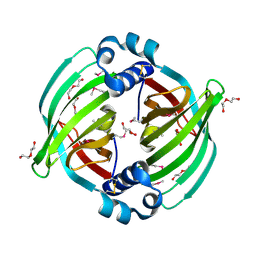 | |
4P6W
 
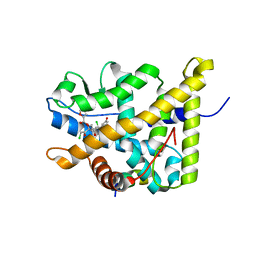 | | Crystal Structure of mometasone furoate-bound glucocorticoid receptor ligand binding domain | | 分子名称: | Glucocorticoid receptor, MOMETASONE FUROATE, Nuclear receptor coactivator 2 | | 著者 | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | 登録日 | 2014-03-25 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
5GZR
 
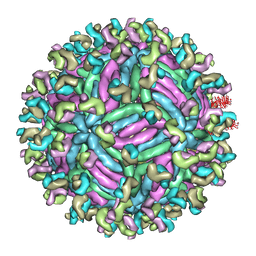 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | 分子名称: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | 著者 | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | 登録日 | 2016-10-01 | | 公開日 | 2016-11-30 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (9.4 Å) | | 主引用文献 | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5Y6M
 
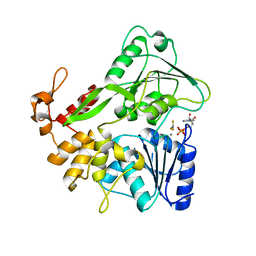 | | Zika virus helicase in complex with ADP-AlF3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase domain from Genome polyprotein, ... | | 著者 | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | 登録日 | 2017-08-12 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
2Q6D
 
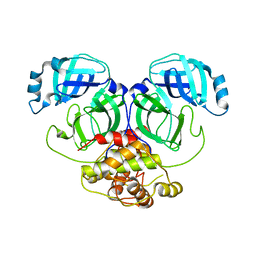 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | 分子名称: | Infectious bronchitis virus (IBV) main protease | | 著者 | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | 登録日 | 2007-06-04 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
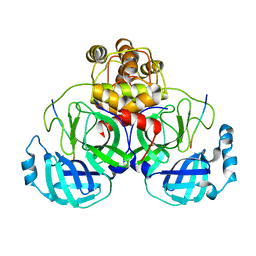 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | 分子名称: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | 著者 | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | 登録日 | 2007-06-05 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | 著者 | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | 登録日 | 2020-03-16 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | 分子名称: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | 登録日 | 2007-06-05 | | 公開日 | 2008-02-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
8H6I
 
 | | The crystal structure of SARS-CoV-2 3C-like protease Double Mutant (L50F and E166V) in complex with a traditional Chinese Medicine Inhibitors | | 分子名称: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H7K
 
 | |
8H7W
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (S144A) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H5F
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) L167F Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H51
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H5P
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
