5KTU
 
 | | Crystal structure of the bromodomain of human CREBBP bound to pyrazolopiperidine scaffold | | 分子名称: | 1-(3-phenylazanyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)ethanone, CREB-binding protein, DIMETHYL SULFOXIDE | | 著者 | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | 登録日 | 2016-07-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | 分子名称: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | 登録日 | 2020-12-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Li, S, Zheng, Q. | | 登録日 | 2019-12-05 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Zheng, Q, Li, S. | | 登録日 | 2019-12-05 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
4FC2
 
 | |
7CHO
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-1D2 with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-1D2 heavy chain, ... | | 著者 | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | 登録日 | 2020-07-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.561 Å) | | 主引用文献 | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHS
 
 | | Crystal structure of SARS-CoV-2 antibody P22A-1D1 with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P22A-1D1 heavy chain, ... | | 著者 | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | 登録日 | 2020-07-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7CHP
 
 | | Crystal structure of SARS-CoV-2 antibody P5A-3C8 with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P5A-3C8 heavy chain, ... | | 著者 | Wang, X, Zhang, L, Ge, J, Wang, R, Zhang, Q. | | 登録日 | 2020-07-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.357 Å) | | 主引用文献 | Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7DPF
 
 | |
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | 分子名称: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Rapp, M, Shapiro, L, Ho, D.D. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
8HK1
 
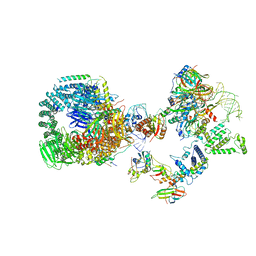 | | The cryo-EM structure of human pre-17S U2 snRNP | | 分子名称: | ATP-dependent RNA helicase DDX42, HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, ... | | 著者 | Zhang, X, Zhan, X, Shi, Y. | | 登録日 | 2022-11-24 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Mechanisms of the RNA helicases DDX42 and DDX46 in human U2 snRNP assembly.
Nat Commun, 14, 2023
|
|
8SMV
 
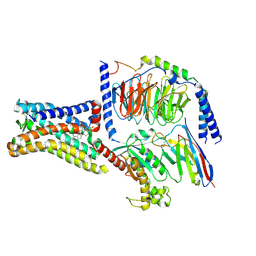 | | GPR161 Gs heterotrimer | | 分子名称: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Hoppe, N, Manglik, A, Harrison, S. | | 登録日 | 2023-04-26 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Biorxiv, 2023
|
|
6R8X
 
 | |
6R9W
 
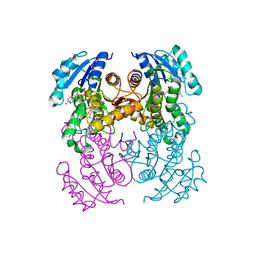 | | Crystal structure of InhA in complex with AP-124 inhibitor | | 分子名称: | (2~{S})-1-(benzimidazol-1-yl)-3-(2,3-dihydro-1~{H}-inden-5-yloxy)propan-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Takebayashi, Y, Hinchliffe, P, Spencer, J. | | 登録日 | 2019-04-04 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Discovery of New and Potent InhA Inhibitors as Antituberculosis Agents: Structure-Based Virtual Screening Validated by Biological Assays and X-ray Crystallography.
J.Chem.Inf.Model., 60, 2020
|
|
8HBM
 
 | |
5E6P
 
 | |
6LBM
 
 | |
6LBI
 
 | |
8H7G
 
 | |
8XC4
 
 | | Nipah virus attachment glycoprotein head domain in complex with a broadly neutralizing antibody 1E5 | | 分子名称: | 1E5-VH, 1E5-VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fan, P.F, Yu, C.M, Chen, W. | | 登録日 | 2023-12-08 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (3.24 Å) | | 主引用文献 | A potent Henipavirus cross-neutralizing antibody reveals a dynamic fusion-triggering pattern of the G-tetramer.
Nat Commun, 15, 2024
|
|
7CQP
 
 | | Complex of TRPC4 and Calmodulin_Nlobe | | 分子名称: | CALCIUM ION, Calmodulin-1, Peptide from Short transient receptor potential channel 4 | | 著者 | Shen, Z.S. | | 登録日 | 2020-08-11 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Calmodulin binds to Drosophila TRP with an unexpected mode.
Structure, 29, 2021
|
|
