6M1V
 
 | |
7FGD
 
 | | A naturally-occurring neuraminidase-inhibitors-resistant NA from asiatic toad influenza B-like virus | | 分子名称: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, toad NA (OSE) | | 著者 | Chai, Y, Wu, Y, Gao, F. | | 登録日 | 2021-07-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7FGC
 
 | |
7FGB
 
 | | A naturally-occurring neuraminidase-inhibitors-resistant NA from asiatic toad influenza B-like virus | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | 著者 | Chai, Y, Wu, Y, Gao, F. | | 登録日 | 2021-07-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and inhibitor sensitivity analysis of influenza B-like viral neuraminidases derived from Asiatic toad and spiny eel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7FGE
 
 | |
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | 著者 | Wu, Y, Qi, J, Gao, F. | | 登録日 | 2020-04-26 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
8XFZ
 
 | | The structure of HLA-A/L1-2 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-14 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKE
 
 | | The structure of HLA-A/14-3-D | | 分子名称: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | 著者 | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | 登録日 | 2023-12-23 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-23 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XG2
 
 | | The structure of HLA-A/Pep14 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-14 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XES
 
 | | The structure of HLA-A/L1-1 | | 分子名称: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | 著者 | Zhang, J.N, Yue, C, Liu, J. | | 登録日 | 2023-12-12 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
5ZL2
 
 | |
5ZKX
 
 | |
7E0B
 
 | |
4F7M
 
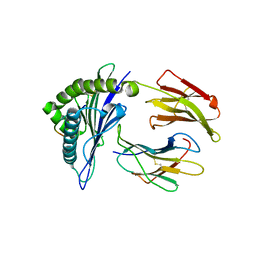 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PA (649-658) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | 著者 | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, George, F.G. | | 登録日 | 2012-05-16 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
8JVA
 
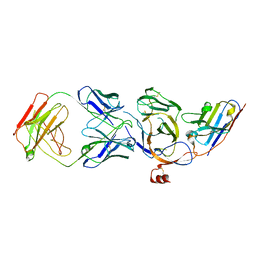 | | Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 heavy chain, S2L20 light chain, ... | | 著者 | Liu, B, Liu, H.H, Han, P, Qi, J.X. | | 登録日 | 2023-06-28 | | 公開日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Enhanced potency of an IgM-like nanobody targeting conserved epitope in SARS-CoV-2 spike N-terminal domain.
Signal Transduct Target Ther, 9, 2024
|
|
8Y0Y
 
 | | Cryo-EM structure of the 123-316 scDb/PT-RBD complex | | 分子名称: | 123-316 scDb, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Jia, G.W, Tong, Z, Tong, J.Y, Su, Z.M. | | 登録日 | 2024-01-23 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
5GZR
 
 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | 分子名称: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | 著者 | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | 登録日 | 2016-10-01 | | 公開日 | 2016-11-30 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (9.4 Å) | | 主引用文献 | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | 著者 | Zhao, R.C, Wu, L.L, Han, P. | | 登録日 | 2022-12-15 | | 公開日 | 2023-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4KPS
 
 | | Structure and receptor binding specificity of the hemagglutinin H13 from avian influenza A virus H13N6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | 著者 | Lu, X, Qi, J, Shi, Y, Gao, G. | | 登録日 | 2013-05-14 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.587 Å) | | 主引用文献 | Structure and Receptor Binding Specificity of Hemagglutinin H13 from Avian Influenza A Virus H13N6
J.Virol., 87, 2013
|
|
4KPQ
 
 | | Structure and receptor binding specificity of the hemagglutinin H13 from avian influenza A virus H13N6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Lu, X, Qi, J, Shi, Y, Gao, G. | | 登録日 | 2013-05-14 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structure and Receptor Binding Specificity of Hemagglutinin H13 from Avian Influenza A Virus H13N6
J.Virol., 87, 2013
|
|
7WBL
 
 | |
7BSD
 
 | |
7BSC
 
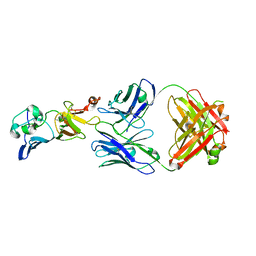 | |
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | 著者 | Zhao, Z, Qi, J, Gao, F.G. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
