8Y4G
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with Bofutrelvir | | 分子名称: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Zeng, P, Wang, W.W, Zhang, J, Li, J. | | 登録日 | 2024-01-30 | | 公開日 | 2025-01-08 | | 最終更新日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8Y4H
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with Bofutrelvir | | 分子名称: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Guo, L, Wang, W.W, Zhang, J, Li, J. | | 登録日 | 2024-01-30 | | 公開日 | 2025-01-08 | | 最終更新日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8YRH
 
 | | Complex of SARS-CoV-2 main protease and Rosmarinic acid | | 分子名称: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, 3C-like proteinase nsp5 | | 著者 | Wang, Q.S, Li, Q.H. | | 登録日 | 2024-03-21 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.841 Å) | | 主引用文献 | Structural basis of rosmarinic acid inhibitory mechanism on SARS-CoV-2 main protease.
Biochem.Biophys.Res.Commun., 724, 2024
|
|
8YWZ
 
 | | Crystal structure of SARS-Cov-2 main protease H163A mutant in complex with Bofutrelvir | | 分子名称: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Zou, X.F, Wang, W.W, Zhang, J, Li, J. | | 登録日 | 2024-04-01 | | 公開日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8Y2H
 
 | |
8YKQ
 
 | |
8YKP
 
 | |
8YKN
 
 | |
8YKO
 
 | |
8YKM
 
 | |
7VR8
 
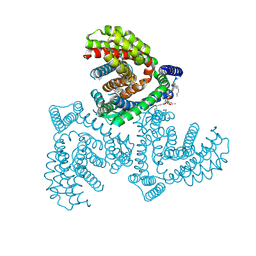 | | Inward-facing structure of human EAAT2 in the substrate-free state | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | 著者 | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2021-10-22 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7VR7
 
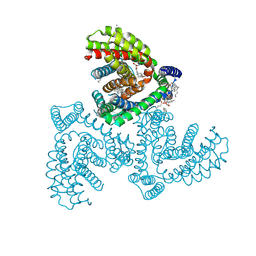 | | Inward-facing structure of human EAAT2 in the WAY213613-bound state | | 分子名称: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2021-10-22 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7WQJ
 
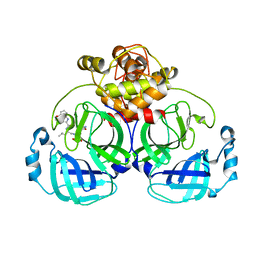 | | Crystal structure of MERS main protease in complex with PF07304814 | | 分子名称: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2022-01-25 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7W9N
 
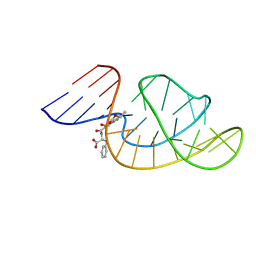 | | THE STRUCTURE OF OBA33-OTA COMPLEX | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, OTA DNA APTAMER (33-MER) | | 著者 | Xu, G.H, Li, C.G. | | 登録日 | 2021-12-10 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insights into the Mechanism of High-Affinity Binding of Ochratoxin A by a DNA Aptamer.
J.Am.Chem.Soc., 144, 2022
|
|
8UPV
 
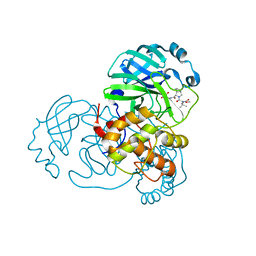 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | 分子名称: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | 著者 | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | 登録日 | 2023-10-23 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | 分子名称: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | 著者 | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | 登録日 | 2023-10-23 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPW
 
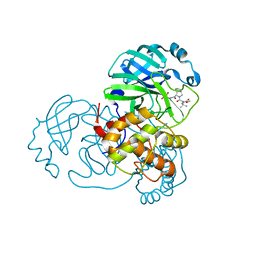 | | Structure of SARS-Cov2 3CLPro in complex with Compound 34 | | 分子名称: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6S)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | 著者 | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | 登録日 | 2023-10-23 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
7XMV
 
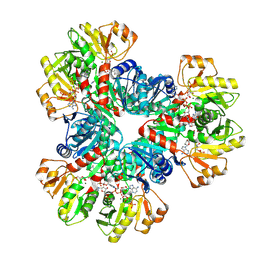 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A(AMP/ADP) filament bound with ADP, AMP and R5P | | 分子名称: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | 登録日 | 2022-04-27 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7XN3
 
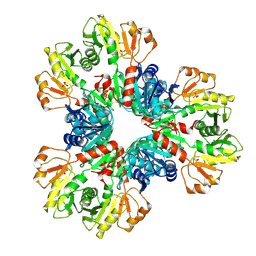 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type B filament bound with Pi | | 分子名称: | PHOSPHATE ION, Ribose-phosphate pyrophosphokinase | | 著者 | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | 登録日 | 2022-04-27 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7XMU
 
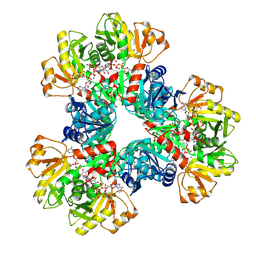 | | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A filament bound with ADP, Pi and R5P | | 分子名称: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Hu, H.H, Lu, G.M, Chang, C.C, Liu, J.L. | | 登録日 | 2022-04-26 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Filamentation modulates allosteric regulation of PRPS.
Elife, 11, 2022
|
|
7DKM
 
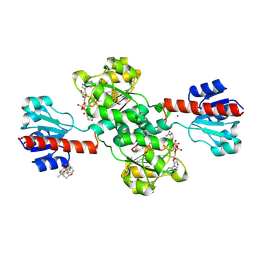 | | PHGDH covalently linked to oridonin | | 分子名称: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, CHLORIDE ION, D-3-phosphoglycerate dehydrogenase, ... | | 著者 | Sun, Q, Lei, Y. | | 登録日 | 2020-11-25 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Biophysical and biochemical properties of PHGDH revealed by studies on PHGDH inhibitors.
Cell.Mol.Life Sci., 79, 2021
|
|
7DFT
 
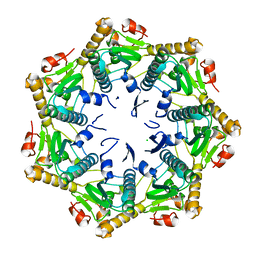 | | Crystal structure of Xanthomonas oryzae ClpP | | 分子名称: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | 著者 | Yang, C.-G, Yang, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Dysregulation of ClpP by Small-Molecule Activators Used Against Xanthomonas oryzae pv. oryzae Infections.
J.Agric.Food Chem., 69, 2021
|
|
7DFU
 
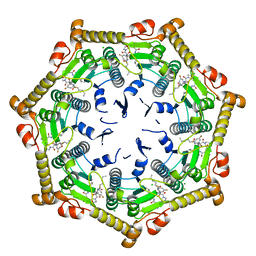 | |
7FIF
 
 | | Cryo-EM structure of the hedgehog release protein Disp from water bear (Hypsibius dujardini) | | 分子名称: | Protein dispatched-like protein 1 | | 著者 | Luo, Y, Wan, G, Wang, Q, Zhao, Y, Cong, Y, Li, D. | | 登録日 | 2021-07-31 | | 公開日 | 2021-09-08 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Architecture of Dispatched, a Transmembrane Protein Responsible for Hedgehog Release.
Front Mol Biosci, 8, 2021
|
|
