6L2H
 
 | | CGTase mutant-Y167H | | 分子名称: | Alpha-cyclodextrin glucanotransferase, CALCIUM ION | | 著者 | Fan, T.W, Hou, A.Q, Chao, Y.P, Sun, Y. | | 登録日 | 2019-10-03 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.096 Å) | | 主引用文献 | Structure basis of a mutant a-CGTase tyrosine167histidine from Bacillus sp. 602-1 with enhanced a-CD production
To Be Published
|
|
2AL4
 
 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH quisqualate and CX614. | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2,3,6A,7,8,9-HEXAHYDRO-11H-[1,4]DIOXINO[2,3-G]PYRROLO[2,1-B][1,3]BENZOXAZIN-11-ONE, Glutamate receptor 2, ... | | 著者 | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | 登録日 | 2005-08-04 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
7DDX
 
 | | Crystal structure of KANK1 S1179F mutant in complex wtih eIF4A1 | | 分子名称: | Eukaryotic initiation factor 4A-I, GLYCEROL, KN motif and ankyrin repeat domains 1, ... | | 著者 | Pan, W, Xu, Y, Wei, Z. | | 登録日 | 2020-10-30 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Nephrotic-syndrome-associated mutation of KANK2 induces pathologic binding competition with physiological interactor KIF21A.
J.Biol.Chem., 297, 2021
|
|
7C7E
 
 | | Crystal structure of C terminal domain of Escherichia coli DgoR | | 分子名称: | Putative DNA-binding transcriptional regulator, SULFATE ION, TRIETHYLENE GLYCOL, ... | | 著者 | Lin, W. | | 登録日 | 2020-05-25 | | 公開日 | 2021-01-20 | | 実験手法 | X-RAY DIFFRACTION (2.047 Å) | | 主引用文献 | Structural and Functional Analyses of the Transcription Repressor DgoR From Escherichia coli Reveal a Divalent Metal-Containing D-Galactonate Binding Pocket.
Front Microbiol, 11, 2020
|
|
7CWS
 
 | |
7CWT
 
 | |
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | 分子名称: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | 著者 | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | 登録日 | 2021-03-23 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG4
 
 | | Cryo-EM structure of nauclefine-induced PDE3A-SLFN12 complex | | 分子名称: | MAGNESIUM ION, Parvine, Schlafen family member 12, ... | | 著者 | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | 登録日 | 2021-03-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG1
 
 | | Cryo-EM structure of DNMDP-induced PDE3A-SLFN12 complex | | 分子名称: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, Schlafen family member 12, ... | | 著者 | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | 登録日 | 2021-03-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7DSS
 
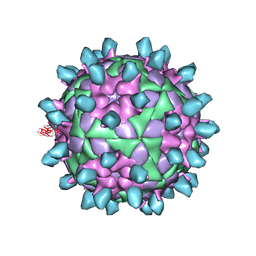 | | Complex of FMDV and M8 Nab | | 分子名称: | M8 Nab, VP1 of O type FMDV capsid, VP2 of O-type FMDV capsid, ... | | 著者 | Dong, H, Liu, P. | | 登録日 | 2021-01-02 | | 公開日 | 2021-03-31 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
7D39
 
 | | FLR-apo | | 分子名称: | Cd1, FLAVIN MONONUCLEOTIDE | | 著者 | Hong, S, Yang, G.H, Zhang, P. | | 登録日 | 2020-09-18 | | 公開日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D38
 
 | | flavone reductase | | 分子名称: | Cd1, FLAVIN MONONUCLEOTIDE, chrysin | | 著者 | Hong, S, Yang, G.H, Zhang, P. | | 登録日 | 2020-09-18 | | 公開日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.649 Å) | | 主引用文献 | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D3B
 
 | | flavone reductase | | 分子名称: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, Cd1, FLAVIN MONONUCLEOTIDE | | 著者 | Hong, S, Yang, G.H, Zhang, P. | | 登録日 | 2020-09-18 | | 公開日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7D3A
 
 | | flavone reductase | | 分子名称: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Cd1, FLAVIN MONONUCLEOTIDE | | 著者 | Hong, S, Yang, G.H, Zhang, P. | | 登録日 | 2020-09-18 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-09-29 | | 実験手法 | X-RAY DIFFRACTION (2.552 Å) | | 主引用文献 | Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria.
Nat Commun, 12, 2021
|
|
7DST
 
 | | FMDV capsid in complex with M170 Nab | | 分子名称: | M170 Nab, VP1 of O type FMDV capsid, VP2 of O type FMDV capsid, ... | | 著者 | Dong, H, Liu, P. | | 登録日 | 2021-01-02 | | 公開日 | 2021-03-10 | | 最終更新日 | 2022-05-25 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
7E7D
 
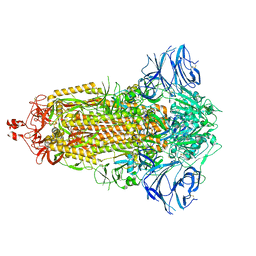 | |
7E7B
 
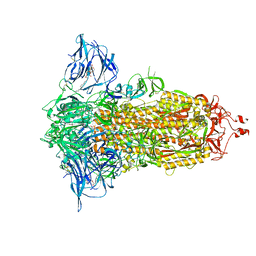 | | Cryo-EM structure of the SARS-CoV-2 furin site mutant S-Trimer from a subunit vaccine candidate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | 著者 | Zheng, S, Ma, J. | | 登録日 | 2021-02-25 | | 公開日 | 2021-03-24 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19.
J.Virol., 95, 2021
|
|
7CWU
 
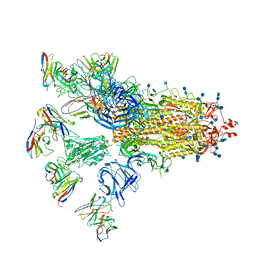 | |
3H6W
 
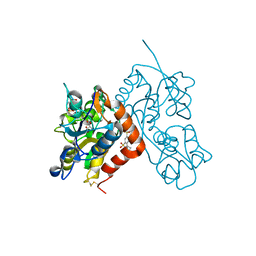 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5217 at 1.50 A resolution | | 分子名称: | (3R)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H6U
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS1493 at 1.85 A resolution | | 分子名称: | (3S)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CITRATE ANION, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | 分子名称: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMR
 
 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
3H6T
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and cyclothiazide at 2.25 A resolution | | 分子名称: | ACETATE ION, CACODYLATE ION, CYCLOTHIAZIDE, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H6V
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution | | 分子名称: | (3R)-3-cyclopentyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | 著者 | Hald, H, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-04-24 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
