8TYZ
 
 | | Structure of a bacterial E1-E2-Ubl complex (form 2) | | 分子名称: | E1(BilD), E2(BilB), Ubl(BilA), ... | | 著者 | Ye, Q, Chambers, L.R, Corbett, K.D. | | 登録日 | 2023-08-26 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | A eukaryotic-like ubiquitination system in bacterial antiviral defence.
Nature, 631, 2024
|
|
5YQX
 
 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | (2R)-2-(cyclopropylmethyl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-4H-1,4-benzoxazin-3-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | 著者 | Xue, X, Zhang, Y, Wang, C, Song, M. | | 登録日 | 2017-11-08 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Benzoxazinone-containing 3,5-dimethylisoxazole derivatives as BET bromodomain inhibitors for treatment of castration-resistant prostate cancer.
Eur.J.Med.Chem., 152, 2018
|
|
4TQD
 
 | | Crystal Structure of the C-terminal domain of IFRS bound with 3-iodo-L-Phe and ATP | | 分子名称: | 1,2-ETHANEDIOL, 3-iodo-L-phenylalanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Nakamura, A, O'Donoghue, P, Soll, D. | | 登録日 | 2014-06-11 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1429 Å) | | 主引用文献 | Polyspecific pyrrolysyl-tRNA synthetases from directed evolution.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8BDP
 
 | |
4IYP
 
 | | structure of the nPP2Ac-alpha4 complex | | 分子名称: | Immunoglobulin-binding protein 1, Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform | | 著者 | Jiang, L, Stanevich, V, Satyshur, K.A, Xing, Y. | | 登録日 | 2013-01-29 | | 公開日 | 2013-04-17 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.797 Å) | | 主引用文献 | Structural basis of protein phosphatase 2A stable latency.
Nat Commun, 4, 2013
|
|
4TQF
 
 | | Crystal Structure of the C-terminal domain of IFRS bound with 2-(5-bromothienyl)-L-Ala and ATP | | 分子名称: | 1,2-ETHANEDIOL, 3-(5-bromothiophen-2-yl)-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Nakamura, A, O'Donoghue, P, Soll, D. | | 登録日 | 2014-06-11 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7143 Å) | | 主引用文献 | Polyspecific pyrrolysyl-tRNA synthetases from directed evolution.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7JHX
 
 | | Crystal structure of hEPG5 LIR/GABARAPL1 complex | | 分子名称: | Ectopic P granules protein 5 homolog, Gamma-aminobutyric acid receptor-associated protein-like 1, SULFATE ION | | 著者 | Cheung, Y.W.S, Yip, C.K. | | 登録日 | 2020-07-21 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Insights on autophagosome-lysosome tethering from structural and biochemical characterization of human autophagy factor EPG5.
Commun Biol, 4, 2021
|
|
8XLV
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-26 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
4UVI
 
 | | Discovery of pyrimidine isoxazoles InhA in complex with compound 23 | | 分子名称: | 5-{[(4,6-dimethylpyrimidin-2-yl)sulfanyl]methyl}-N-[(2-methylpyridin-4-yl)methyl]-1,2-oxazole-3-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Read, J.A, Gingell, H, Madhavapeddi, P, Ghorpade, S, Cowan, S. | | 登録日 | 2014-08-05 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Hitting the Target in More Than One Way: Novel, Direct Inhibitors of Mycobacterium Tuberculosis Enoyl Acp Reductase
To be Published
|
|
4UVD
 
 | | Discovery of pyrimidine isoxazoles InhA in complex with compound 6 | | 分子名称: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[(2Z)-5-[3-(trifluoromethyl)benzyl]-1,3-thiazol-2(3H)-ylidene]acetamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], MAGNESIUM ION, ... | | 著者 | Read, J.A, Gingell, H, Madhavapeddi, P, Ghorpade, S, Cowan, S. | | 登録日 | 2014-08-05 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Hitting the Target in More Than One Way: Novel, Direct Inhibitors of Mycobacterium Tuberculosis Enoyl Acp Reductase
To be Published
|
|
8C8G
 
 | |
8XMT
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P), RBD-closed state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-28 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P) in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-29 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN3
 
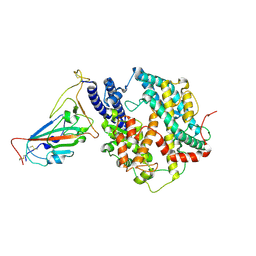 | | SARS-CoV-2 Omicron HV.1 RBD in complex with human ACE2 (local refinement from the spike protein) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-28 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XNK
 
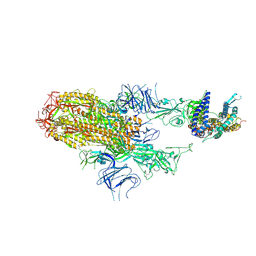 | | Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P) in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-30 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (2.78 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XM5
 
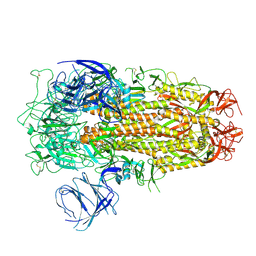 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-27 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XMG
 
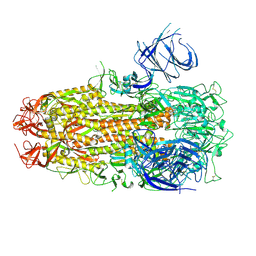 | | Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P), RBD-closed state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-27 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XNF
 
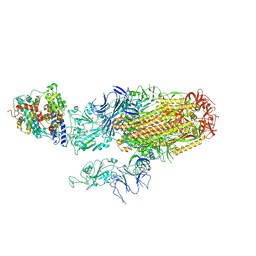 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-29 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN2
 
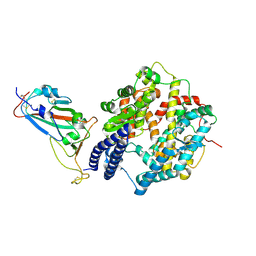 | | SARS-CoV-2 Omicron EG.5.1 RBD in complex with human ACE2 (local refined from the spike protein) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | 登録日 | 2023-12-28 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y5J
 
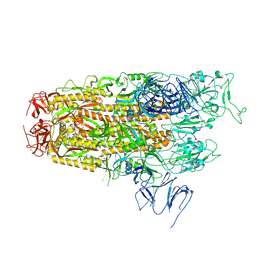 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | 登録日 | 2024-01-31 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
3OHI
 
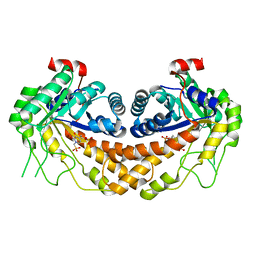 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with 3-hydroxy-2-pyridone | | 分子名称: | ({3-hydroxy-2-oxo-4-[2-(phosphonooxy)ethyl]pyridin-1(2H)-yl}methyl)phosphonic acid, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | 著者 | Herzberg, O, Galkin, A. | | 登録日 | 2010-08-17 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Rational design, synthesis and evaluation of first generation inhibitors of the Giardia lamblia fructose-1,6-biphosphate aldolase.
J.Inorg.Biochem., 105, 2010
|
|
8QEO
 
 | |
8QEN
 
 | |
7CGG
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | 分子名称: | CHLORIDE ION, PBE-5A (5'-R(P*UP*GP*UP*AP*AP*AP*UP*A)-3'), PUM-HD domain-containing protein | | 著者 | Zheng, X, Yunyu, S. | | 登録日 | 2020-07-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGF
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | 分子名称: | CHLORIDE ION, PBE-5G (5'-R(P*UP*GP*UP*AP*GP*AP*UP*A)-3'), PUM-HD domain-containing protein | | 著者 | Zheng, X, Yunyu, S. | | 登録日 | 2020-07-01 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
