6JHV
 
 | |
5UC0
 
 | | Crystal Structure of Beta-barrel-like, Uncharacterized Protein of COG5400 from Brucella abortus | | 分子名称: | CHLORIDE ION, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | 著者 | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-12-21 | | 公開日 | 2017-02-08 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Periplasmic protein EipA determines envelope stress resistance and virulence in Brucella abortus.
Mol. Microbiol., 2018
|
|
8CP7
 
 | |
3GFV
 
 | | Crystal Structure of Petrobactin-binding Protein YclQ from Bacillu subtilis | | 分子名称: | ASPARAGINE, PHOSPHATE ION, Uncharacterized ABC transporter solute-binding protein yclQ | | 著者 | Kim, Y, Maltseva, N, Zawadzka, A.M, Raymond, K.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-02-27 | | 公開日 | 2009-05-12 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Characterization of a Bacillus subtilis transporter for petrobactin, an anthrax stealth siderophore.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7F4U
 
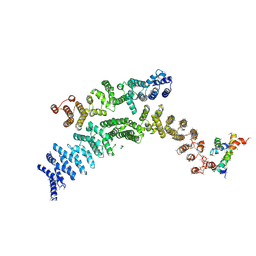 | | Cryo-EM structure of TELO2-TTI1-TTI2 complex | | 分子名称: | TELO2-interacting protein 1 homolog, TELO2-interacting protein 2, Telomere length regulation protein TEL2 homolog | | 著者 | Cho, Y, Kim, Y. | | 登録日 | 2021-06-21 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structure of the Human TELO2-TTI1-TTI2 Complex.
J.Mol.Biol., 434, 2022
|
|
7RXU
 
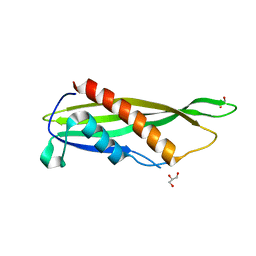 | | Crystal structure of Cj1090c | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Lipoprotein | | 著者 | Kim, Y, Yeo, H.J. | | 登録日 | 2021-08-23 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of Campylobacter jejuni lipoprotein Cj1090c.
Proteins, 91, 2023
|
|
6VXS
 
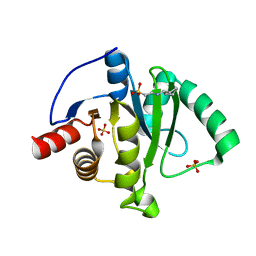 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Non-structural protein 3, ... | | 著者 | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Mesecar, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-24 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
6W61
 
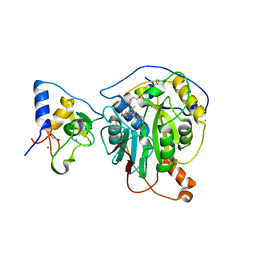 | | Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2. | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase, CHLORIDE ION, ... | | 著者 | Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine
Biorxiv, 2020
|
|
6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | 分子名称: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | 著者 | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-03-04 | | 公開日 | 2020-03-25 | | 最終更新日 | 2022-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6W08
 
 | | Crystal Structure of Motility Associated Killing Factor E from Vibrio cholerae | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | 著者 | Kim, Y, Jedrzejczak, R, Joachimiak, G, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-29 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6VYO
 
 | | Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | 著者 | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-27 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6WKP
 
 | | Crystal structure of RNA-binding domain of nucleocapsid phosphoprotein from SARS CoV-2, monoclinic crystal form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | 著者 | Chang, C, Michalska, K, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-16 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
4ZWV
 
 | | Crystal Structure of Aminotransferase AtmS13 from Actinomadura melliaura | | 分子名称: | GLYCEROL, Putative aminotransferase | | 著者 | Kim, Y, Bigelow, L, Endres, M, Wang, F, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-19 | | 公開日 | 2015-06-03 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
6WT2
 
 | | Crystal Structure of Putative NAD(P)H-Flavin Oxidoreductase from Neisseria meningitidis | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-01 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
1MKI
 
 | | Crystal Structure of Bacillus Subtilis Probable Glutaminase, APC1040 | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, Probable Glutaminase ybgJ | | 著者 | Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-08-29 | | 公開日 | 2003-06-03 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
4I4K
 
 | | Streptomyces globisporus C-1027 9-membered enediyne conserved protein SgcE6 | | 分子名称: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | 著者 | Kim, Y, Bigelow, L, Clancy, S, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-11-27 | | 公開日 | 2012-12-12 | | 最終更新日 | 2016-12-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J Antibiot (Tokyo), 69, 2016
|
|
4IXH
 
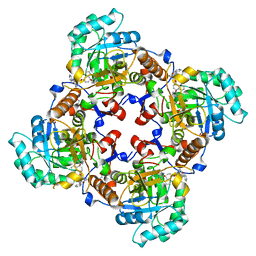 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Cryptosporidium parvum | | 分子名称: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, INOSINIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-01-25 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.105 Å) | | 主引用文献 | Optimization of Benzoxazole-Based Inhibitors of Cryptosporidium parvum Inosine 5'-Monophosphate Dehydrogenase.
J.Med.Chem., 56, 2013
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | 分子名称: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | 著者 | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-11-27 | | 公開日 | 2012-12-12 | | 最終更新日 | 2022-05-04 | | 実験手法 | X-RAY DIFFRACTION (2.784 Å) | | 主引用文献 | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
3DX5
 
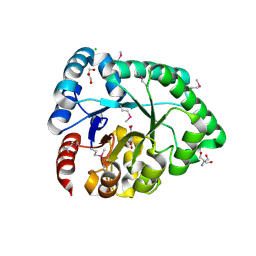 | | Crystal structure of the probable 3-DHS dehydratase AsbF involved in the petrobactin synthesis from Bacillus anthracis | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, ... | | 著者 | Kim, Y, Maltseva, N, Stols, L, Eschenfeldt, W, Pfleger, B.F, Sherman, D.H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-07-23 | | 公開日 | 2008-09-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structural and functional analysis of AsbF: origin of the stealth 3,4-dihydroxybenzoic acid subunit for petrobactin biosynthesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3USB
 
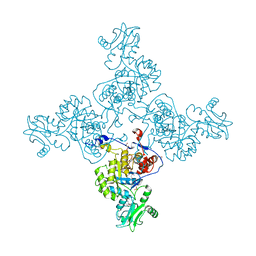 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | 分子名称: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-11-23 | | 公開日 | 2011-12-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
6NTR
 
 | | Crystal Structure of Beta-barrel-like Protein of Domain of Unknown Function DUF1849 from Brucella abortus | | 分子名称: | 1,2-ETHANEDIOL, ATP/GTP-binding site-containing protein A, GLYCEROL | | 著者 | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2019-01-30 | | 公開日 | 2019-02-13 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | BrucellaPeriplasmic Protein EipB Is a Molecular Determinant of Cell Envelope Integrity and Virulence.
J.Bacteriol., 201, 2019
|
|
2H7H
 
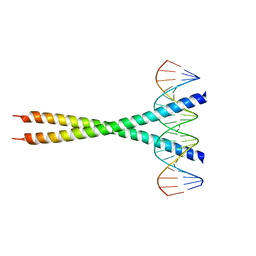 | |
4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | 分子名称: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | 著者 | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-07-25 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.902 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3TSD
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | 分子名称: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-13 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.653 Å) | | 主引用文献 | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TSB
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | 分子名称: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | 著者 | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-12 | | 公開日 | 2011-10-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.595 Å) | | 主引用文献 | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
