8IOS
 
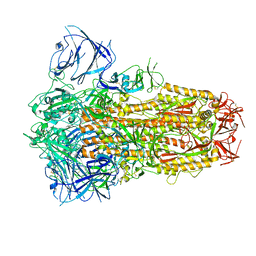 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-1 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOU
 
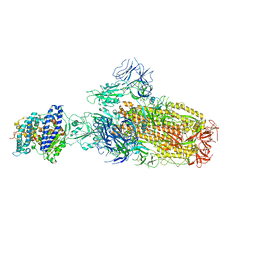 | | Structure of SARS-CoV-2 XBB.1 spike glycoprotein in complex with ACE2 (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOV
 
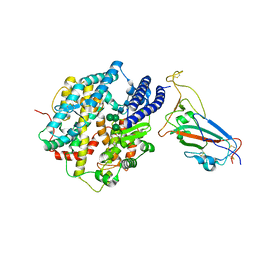 | | Structure of SARS-CoV-2 XBB.1 spike RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOT
 
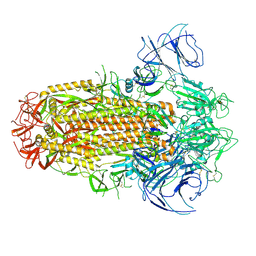 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-2 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-15 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-15 | | 公開日 | 2023-10-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | 著者 | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-11-08 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8K5H
 
 | | Structure of the SARS-CoV-2 BA.1 spike with UT28-RD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | 登録日 | 2023-07-21 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Rational in silico design identifies two mutations that restore UT28K SARS-CoV-2 monoclonal antibody activity against Omicron BA.1.
Structure, 32, 2024
|
|
8K5G
 
 | | Structure of the SARS-CoV-2 BA.1 RBD with UT28-RD | | 分子名称: | Spike protein S1, UT28K-RD Fab Heavy chain, UT28K-RD Fab Light chain | | 著者 | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | 登録日 | 2023-07-21 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Rational in silico design identifies two mutations that restore UT28K SARS-CoV-2 monoclonal antibody activity against Omicron BA.1.
Structure, 32, 2024
|
|
7XBD
 
 | | Cryo-EM structure of human galanin receptor 2 | | 分子名称: | Galanin, Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Ishimoto, N, Kita, S, Park, S.Y. | | 登録日 | 2022-03-21 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structure of the human galanin receptor 2 bound to galanin and Gq reveals the basis of ligand specificity and how binding affects the G-protein interface.
Plos Biol., 20, 2022
|
|
6K6N
 
 | | Crystal structure of SIVmac239 Nef protein | | 分子名称: | Protein Nef | | 著者 | Hirao, K, Andrews, S, Kuroki, K, Kusaka, H, Tadokoro, T, Kita, S, Ose, T, Rowland-Jones, S, Maenaka, K. | | 登録日 | 2019-06-04 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.0002 Å) | | 主引用文献 | Structure of HIV-2 Nef Reveals Features Distinct from HIV-1 Involved in Immune Regulation.
Iscience, 23, 2020
|
|
6K6M
 
 | | Crystal structure of HIV-2 Nef protein | | 分子名称: | Protein Nef | | 著者 | Hirao, K, Andrews, S, Kuroki, K, Kusaka, H, Tadokoro, T, Kita, S, Ose, T, Rowland-Jones, S, Maenaka, K. | | 登録日 | 2019-06-04 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.072 Å) | | 主引用文献 | Structure of HIV-2 Nef Reveals Features Distinct from HIV-1 Involved in Immune Regulation.
Iscience, 23, 2020
|
|
5YCQ
 
 | | Unique Specificity-Enhancing Factor for the AAA+ Lon Protease | | 分子名称: | Heat shock protein HspQ | | 著者 | Abe, Y, Shioi, S, Kita, S, Nakata, H, Maenaka, K, Kohda, D, Katayama, T, Ueda, T. | | 登録日 | 2017-09-08 | | 公開日 | 2018-04-11 | | 実験手法 | X-RAY DIFFRACTION (2.503 Å) | | 主引用文献 | X-ray crystal structure of Escherichia coli HspQ, a protein involved in the retardation of replication initiation
FEBS Lett., 591, 2017
|
|
6K60
 
 | | Structural and functional basis for HLA-G isoform recognition of immune checkpoint receptor LILRBs | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain G, ... | | 著者 | Kuroki, K, Matsubara, H, Kanda, R, Miyashita, N, Shiroishi, M, Fukunaga, Y, Kamishikiryo, J, Fukunaga, A, Hirose, K, Sugita, Y, Kita, S, Ose, T, Maenaka, K. | | 登録日 | 2019-05-31 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.149 Å) | | 主引用文献 | Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms.
J Immunol., 203, 2019
|
|
4W96
 
 | | Crystal structure of cross-linked tetragonal hen egg white lysozyme soaked with 5mM [Ru(CO)3Cl2]2 followed by the reaction in deoxy-myoglobin solution | | 分子名称: | CHLORIDE ION, DIMETHYLFORMAMIDE, Lysozyme C, ... | | 著者 | Tabe, H, Fujita, K, Abe, S, Tsujimoto, M, Kuchimaru, T, Kizaka-Kondo, S, Takano, M, Kitagawa, S, Ueno, T. | | 登録日 | 2014-08-27 | | 公開日 | 2014-12-31 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold
Inorg.Chem., 54, 2015
|
|
1HR9
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Malate Dehydrogenase Signal Peptide | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALATE DEHYDROGENASE, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | 著者 | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | 登録日 | 2000-12-21 | | 公開日 | 2001-07-11 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
4W94
 
 | | Crystal structure of cross-linked tetragonal hen egg white lysozyme soaked with 5mM [Ru(CO)3Cl2]2 | | 分子名称: | CHLORIDE ION, DIMETHYLFORMAMIDE, Lysozyme C, ... | | 著者 | Tabe, H, Fujita, K, Abe, S, Tsujimoto, M, Kuchimaru, T, Kizaka-Kondo, S, Takano, M, Kitagawa, S, Ueno, T. | | 登録日 | 2014-08-27 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold
Inorg.Chem., 54, 2015
|
|
1J19
 
 | | Crystal structure of the radxin FERM domain complexed with the ICAM-2 cytoplasmic peptide | | 分子名称: | 16-mer peptide from Intercellular adhesion molecule-2, radixin | | 著者 | Hamada, K, Shimizu, T, Yonemura, S, Tsukita, S, Tsukita, S, Hakoshima, T. | | 登録日 | 2002-12-02 | | 公開日 | 2003-03-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex
EMBO J., 22, 2003
|
|
1GC6
 
 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN COMPLEXED WITH INOSITOL-(1,4,5)-TRIPHOSPHATE | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, RADIXIN | | 著者 | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | 登録日 | 2000-07-21 | | 公開日 | 2000-09-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
1GC7
 
 | | CRYSTAL STRUCTURE OF THE RADIXIN FERM DOMAIN | | 分子名称: | RADIXIN | | 著者 | Hamada, K, Shimizu, T, Matsui, T, Tsukita, S, Tsukita, S, Hakoshima, T. | | 登録日 | 2000-07-21 | | 公開日 | 2000-09-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain.
EMBO J., 19, 2000
|
|
6YAA
 
 | | Structure of the (SR) Ca2+-ATPase bound to the inhibitor compound CAD204520 and TNP-ATP | | 分子名称: | 4-[2-[(2~{R})-2-[3-propyl-6-(trifluoromethyloxy)-1~{H}-indol-2-yl]piperidin-1-yl]ethyl]morpholine, POTASSIUM ION, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE, ... | | 著者 | Heit, S, Marchesini, M, Gherli, A, Montanaro, A, Patrizi, L, Sorrentino, C, Pagliaro, L, Rompietti, C, Kitara, S, Olesen, C.E, Moller, J.V, Savi, M, Bocchi, L, Vilella, R, Rizzi, F, Baglione, M, Rastelli, G, Loiacona, C, La Starza, R, Mecucci, C, Stegmair, K, Aversa, F, Stilli, D, Lund Winther, A.M, Sportoletti, P, Dalby-Brown, W, Roti, G, Bublitz, M. | | 登録日 | 2020-03-11 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia.
Cell Chem Biol, 27, 2020
|
|
6E69
 
 | | Ortho-substituted phenyl sulfonyl fluoride and fluorosulfate as potent elastase inhibitory fragments | | 分子名称: | 2-(fluorosulfonyl)benzene-1-sulfonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wolan, D.W, Woehl, J.L, Kitamura, S. | | 登録日 | 2018-07-24 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | SuFEx-enabled, agnostic discovery of covalent inhibitors of human neutrophil elastase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1ISN
 
 | | Crystal structure of merlin FERM domain | | 分子名称: | merlin | | 著者 | Shimizu, T, Seto, A, Maita, N, Hamada, K, Tsukita, S, Tsukita, S, Hakoshima, T. | | 登録日 | 2001-12-13 | | 公開日 | 2002-04-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain.
J.Biol.Chem., 277, 2002
|
|
6UKD
 
 | |
4J7V
 
 | | Crystal structure of cross-linked hen egg white lysozyme soaked with 5mM [Ru(benzene)Cl2]2 | | 分子名称: | Benzeneruthenium(II) chloride, CHLORIDE ION, Lysozyme C, ... | | 著者 | Tabe, H, Abe, S, Hikage, T, Kitagawa, S, Ueno, T. | | 登録日 | 2013-02-14 | | 公開日 | 2014-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Solid Artificial Metalloenzymes: Post-Engineering of Porous Protein Crystals by Organometallic Complexes
To be Published
|
|
