8TUK
 
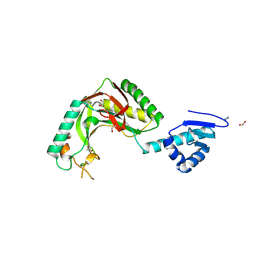 | | Alvinella ASCC1 KH and Phosphodiesterase/Ligase Domain | | 分子名称: | 1,2-ETHANEDIOL, Activating signal cointegrator 1 complex subunit 1, IMIDAZOLE | | 著者 | Tsutakawa, S.E, Tainer, J.A, Arvai, A.S, Chinnam, N.B. | | 登録日 | 2023-08-16 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | ASCC1 structures and bioinformatics reveal a novel helix-clasp-helix RNA-binding motif linked to a two-histidine phosphodiesterase.
J.Biol.Chem., 300, 2024
|
|
8TLY
 
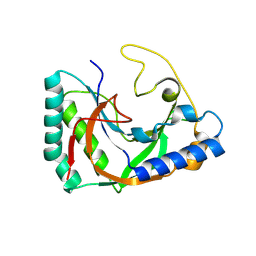 | |
5K8D
 
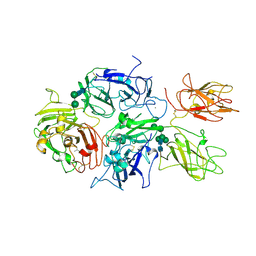 | | Crystal structure of rFVIIIFc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | 著者 | Leksa, N, Quan, C. | | 登録日 | 2016-05-29 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (4.19 Å) | | 主引用文献 | The structural basis for the functional comparability of factor VIII and the long-acting variant recombinant factor VIII Fc fusion protein.
J. Thromb. Haemost., 15, 2017
|
|
6P7M
 
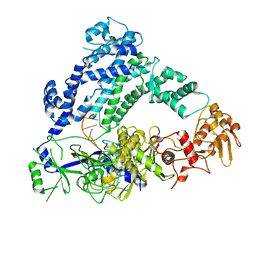 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex) | | 分子名称: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | 著者 | Knott, G.J, Liu, J.J, Doudna, J.A. | | 登録日 | 2019-06-06 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
6P7N
 
 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (2:2 complex) | | 分子名称: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | 著者 | Knott, G.J, Liu, J.J, Doudna, J.A. | | 登録日 | 2019-06-06 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
7T1J
 
 | | Crystal structure of RUBISCO from Rhodospirillaceae bacterium BRH_c57 | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | 著者 | Pereira, J.H, Liu, A.K, Shih, P.M, Adams, P.D. | | 登録日 | 2021-12-02 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural plasticity enables evolution and innovation of RuBisCO assemblies.
Sci Adv, 8, 2022
|
|
7T1C
 
 | |
6DCX
 
 | |
7MZT
 
 | |
8U66
 
 | | Firmicutes Rubisco | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Rubisco | | 著者 | Kaeser, B.P, Liu, A.K, Shih, P.M. | | 登録日 | 2023-09-13 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.21 Å) | | 主引用文献 | Deep-branching evolutionary intermediates reveal structural origins of form I rubisco.
Curr.Biol., 33, 2023
|
|
7WVH
 
 | |
4F52
 
 | | Structure of a Glomulin-RBX1-CUL1 complex | | 分子名称: | Cullin-1, E3 ubiquitin-protein ligase RBX1, Glomulin, ... | | 著者 | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | 登録日 | 2012-05-11 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of a Glomulin-RBX1-CUL1 Complex: Inhibition of a RING E3 Ligase through Masking of Its E2-Binding Surface.
Mol.Cell, 47, 2012
|
|
6WIQ
 
 | | Crystal structure of the co-factor complex of NSP7 and the C-terminal domain of NSP8 from SARS CoV-2 | | 分子名称: | Non-structural protein 7, Non-structural protein 8 | | 著者 | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6WQD
 
 | | The 1.95 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | 著者 | Kim, Y, Wilamowski, M, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-28 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | 著者 | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-20 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
8D3T
 
 | | Crystal structure of GalS1 from Populus trichocarpas | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Pereira, J.H, Prabhakar, P.K, Urbanowicz, B.R, Adams, P.D. | | 登録日 | 2022-06-01 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-04-05 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural and biochemical insight into a modular beta-1,4-galactan synthase in plants.
Nat.Plants, 9, 2023
|
|
8D3Z
 
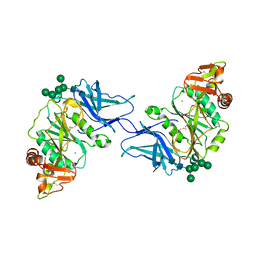 | | Crystal structure of GalS1 in complex with Manganese from Populus trichocarpas | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Galactan synthase, ... | | 著者 | Pereira, J.H, Prabhakar, P.K, Urbanowicz, B.R, Adams, P.D. | | 登録日 | 2022-06-01 | | 公開日 | 2023-03-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural and biochemical insight into a modular beta-1,4-galactan synthase in plants.
Nat.Plants, 9, 2023
|
|
5I99
 
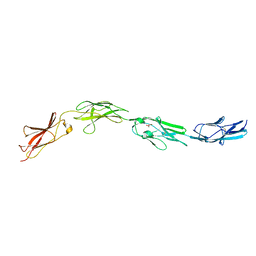 | |
4LCD
 
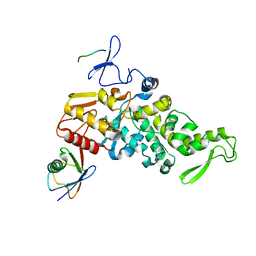 | |
5IKN
 
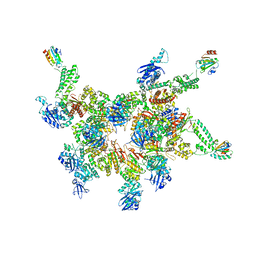 | |
5ZXV
 
 | |
6URA
 
 | | Crystal structure of RUBISCO from Promineofilum breve | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain | | 著者 | Pereira, J.H, Banda, D.M, Liu, A.K, Shih, P.M, Adams, P.D. | | 登録日 | 2019-10-23 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Novel bacterial clade reveals origin of form I Rubisco.
Nat.Plants, 6, 2020
|
|
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | 著者 | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | 登録日 | 2017-12-08 | | 公開日 | 2018-08-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
6VBH
 
 | | Human XPG endonuclease catalytic domain | | 分子名称: | DNA repair protein complementing XP-G cells,Flap endonuclease 1, SULFATE ION | | 著者 | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | 登録日 | 2019-12-18 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.995 Å) | | 主引用文献 | Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7SD1
 
 | |
