6CUF
 
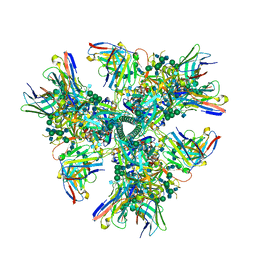 | | Cryo-EM structure at 4.2 A resolution of vaccine-elicited antibody vFP1.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Acharya, P, Carragher, B, Potter, C.S, Kwong, P.D. | | 登録日 | 2018-03-26 | | 公開日 | 2018-07-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Complete functional mapping of infection- and vaccine-elicited antibodies against the fusion peptide of HIV.
PLoS Pathog., 14, 2018
|
|
6CUE
 
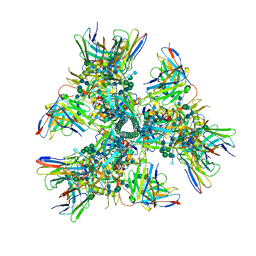 | | Cryo-EM structure at 4.0 A resolution of vaccine-elicited antibody vFP7.04 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Acharya, P, Carragher, B, Potter, C.S, Kwong, P.D. | | 登録日 | 2018-03-26 | | 公開日 | 2018-07-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Complete functional mapping of infection- and vaccine-elicited antibodies against the fusion peptide of HIV.
PLoS Pathog., 14, 2018
|
|
6UKJ
 
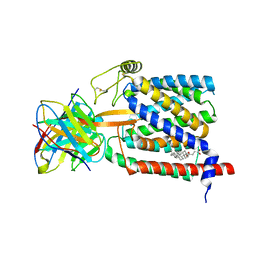 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | 著者 | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | 登録日 | 2019-10-05 | | 公開日 | 2019-12-04 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
6UPL
 
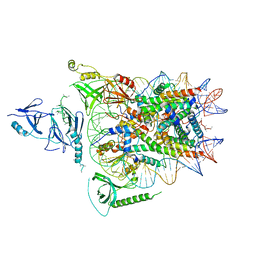 | | Structure of FACT_subnucleosome complex 2 | | 分子名称: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | 著者 | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | 登録日 | 2019-10-17 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
6UPK
 
 | | Structure of FACT_subnucleosome complex 1 | | 分子名称: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | 著者 | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | 登録日 | 2019-10-17 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
6BDF
 
 | | 2.8 A resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy | | 分子名称: | Proteasome subunit alpha, Proteasome subunit beta | | 著者 | Campbell, M.G, Veesler, D, Cheng, A, Potter, C.S, Carragher, B. | | 登録日 | 2017-10-23 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | 2.8 angstrom resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy.
Elife, 4, 2015
|
|
6CE9
 
 | | Insulin Receptor ectodomain in complex with two insulin molecules | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G, Dandey, V.P, Zhang, Z, Strickland, C, Potter, C.S, Carragher, B. | | 登録日 | 2018-02-11 | | 公開日 | 2018-03-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis.
Nature, 556, 2018
|
|
6CEB
 
 | | Insulin Receptor ectodomain in complex with two insulin molecules - C1 symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G, Dandey, V.P, Zhang, Z, Strickland, C, Potter, C.S, Carragher, B. | | 登録日 | 2018-02-11 | | 公開日 | 2018-03-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis.
Nature, 556, 2018
|
|
6CE7
 
 | | Insulin Receptor ectodomain in complex with one insulin molecule | | 分子名称: | Insulin A chain, Insulin B chain, Insulin receptor, ... | | 著者 | Scapin, G, Dandey, V.P, Zhang, Z, Strickland, C, Potter, C.S, Carragher, B. | | 登録日 | 2018-02-11 | | 公開日 | 2018-03-14 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis.
Nature, 556, 2018
|
|
8TOE
 
 | |
8TO6
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter | | 分子名称: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Darst, S.A, Saecker, R.M, Mueller, A.U. | | 登録日 | 2023-08-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TOM
 
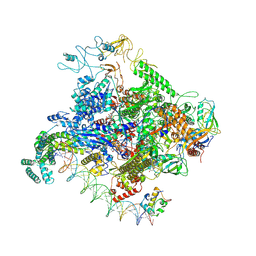 | |
8TO8
 
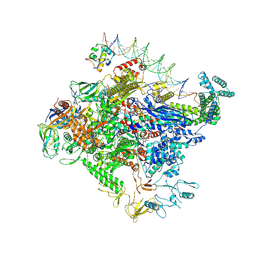 | | Escherichia coli RNA polymerase unwinding intermediate (I1b) at the lambda PR promoter | | 分子名称: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Darst, S.A, Saecker, R.M, Mueller, A.U. | | 登録日 | 2023-08-03 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
8TO1
 
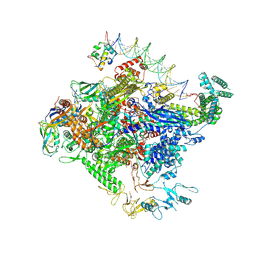 | | Escherichia coli RNA polymerase unwinding intermediate (I1a) at the lambda PR promoter | | 分子名称: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Darst, S.A, Saecker, R.M, Mueller, A.U. | | 登録日 | 2023-08-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
5WOB
 
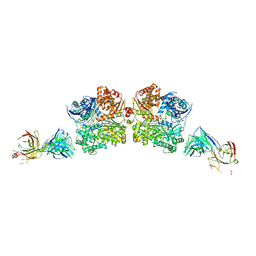 | | Crystal Structure Analysis of Fab1-Bound Human Insulin Degrading Enzyme (IDE) in Complex with Insulin | | 分子名称: | IDE-bound Fab heavy chain, IDE-bound Fab light chain, Insulin, ... | | 著者 | McCord, L.A, Liang, W.G, Farcasanu, M, Wang, A.G, Koide, S, Tang, W.J. | | 登録日 | 2017-08-01 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.95 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6OT1
 
 | |
6OSY
 
 | |
3J5M
 
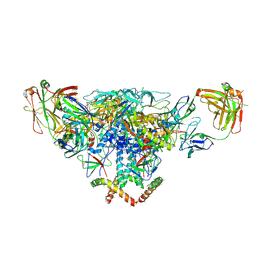 | | Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 3 PGV04 Fabs | | 分子名称: | BG505 SOSIP gp120, BG505 SOSIP gp41, PGV04 heavy chain, ... | | 著者 | Lyumkis, D, Julien, J.-P, Wilson, I.A, Ward, A.B. | | 登録日 | 2013-10-26 | | 公開日 | 2013-11-13 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
5TKJ
 
 | | Structure of vaccine-elicited diverse HIV-1 neutralizing antibody vFP1.01 in complex with HIV-1 fusion peptide residue 512-519 | | 分子名称: | HIV-1 fusion peptide residue 512-519, SULFATE ION, vFP1.01 chimeric mouse antibody heavy chain, ... | | 著者 | Xu, K, Liu, K, Kwong, P.D. | | 登録日 | 2016-10-06 | | 公開日 | 2018-04-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.118 Å) | | 主引用文献 | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
5TKK
 
 | |
4O9U
 
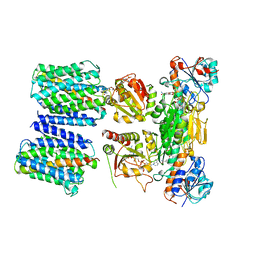 | | Mechanism of transhydrogenase coupling proton translocation and hydride transfer | | 分子名称: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta, NAD/NADP transhydrogenase alpha subunit 1, ... | | 著者 | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | 登録日 | 2014-01-02 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (6.926 Å) | | 主引用文献 | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
4O9P
 
 | | Crystal structure of Thermus thermophilis transhydrogeanse domain II dimer SeMet derivative | | 分子名称: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta | | 著者 | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | 登録日 | 2014-01-02 | | 公開日 | 2014-06-11 | | 最終更新日 | 2015-01-28 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
6E6B
 
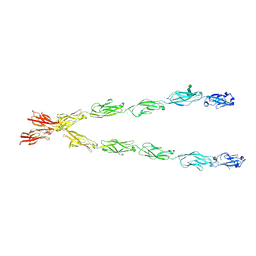 | | Crystal structure of the Protocadherin GammaB4 extracellular domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | 登録日 | 2018-07-24 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.52 Å) | | 主引用文献 | Visualization of clustered protocadherin neuronal self-recognition complexes.
Nature, 569, 2019
|
|
8G9B
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; compressed filament segment reconstruction | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | 著者 | O'Neill, A.G, Kollman, J.M. | | 登録日 | 2023-02-21 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8G8F
 
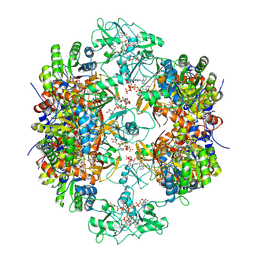 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | 著者 | O'Neill, A.G, Kollman, J.M. | | 登録日 | 2023-02-17 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
