5VN8
 
 | | Cryo-EM model of B41 SOSIP.664 in complex with fragment antigen binding variable domain of b12 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | 著者 | Ozorowski, G, Pallesen, J, Ward, A.B, Cottrell, C.A. | | 登録日 | 2017-04-28 | | 公開日 | 2017-07-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike.
Nature, 547, 2017
|
|
4PMW
 
 | |
5VN3
 
 | | Cryo-EM model of B41 SOSIP.664 in complex with soluble CD4 (D1-D2) and fragment antigen binding variable domain of 17b | | 分子名称: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ozorowski, G, Pallesen, J, Ward, A.B. | | 登録日 | 2017-04-28 | | 公開日 | 2017-07-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike.
Nature, 547, 2017
|
|
6CS2
 
 | | SARS Spike Glycoprotein - human ACE2 complex, Stabilized variant, all ACE2-bound particles | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CS1
 
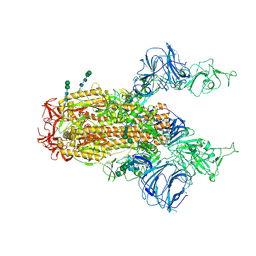 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRX
 
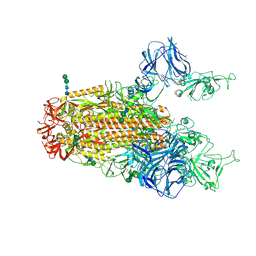 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6DJB
 
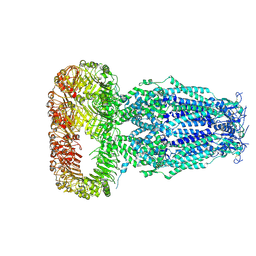 | | Structure of human Volume Regulated Anion Channel composed of SWELL1 (LRRC8A) | | 分子名称: | Volume-regulated anion channel subunit LRRC8A | | 著者 | Kefauver, J.M, Saotome, K, Pallesen, J, Cottrell, C.A, Ward, A.B, Patapoutian, A. | | 登録日 | 2018-05-24 | | 公開日 | 2018-08-15 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structure of the human volume regulated anion channel.
Elife, 7, 2018
|
|
6PZ8
 
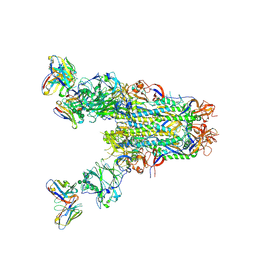 | | MERS S0 trimer in complex with variable domain of antibody G2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G2 heavy chain, ... | | 著者 | Bowman, C.A, Pallesen, J, Ward, A.B. | | 登録日 | 2019-07-31 | | 公開日 | 2019-10-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.19 Å) | | 主引用文献 | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
6CRW
 
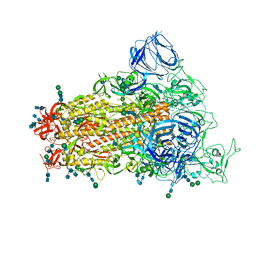 | | SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
5W0B
 
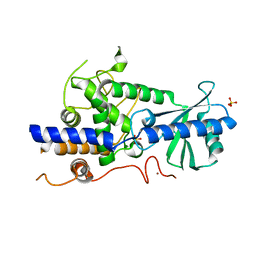 | | Structure of human TUT7 catalytic module (CM) | | 分子名称: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | 著者 | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | 登録日 | 2017-05-30 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.614 Å) | | 主引用文献 | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5W0O
 
 | |
5W0N
 
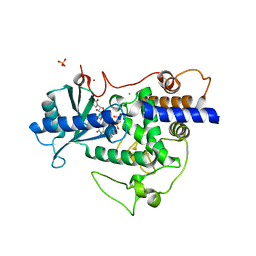 | | Structure of human TUT7 catalytic module (CM) in complex with UMPNPP and U2 RNA | | 分子名称: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, IODIDE ION, MAGNESIUM ION, ... | | 著者 | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | 登録日 | 2017-05-31 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.497 Å) | | 主引用文献 | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5W0M
 
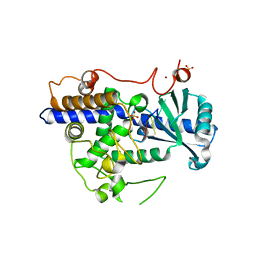 | | Structure of human TUT7 catalytic module (CM) in complex with U5 RNA | | 分子名称: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | 著者 | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | 登録日 | 2017-05-31 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.298 Å) | | 主引用文献 | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6FZ4
 
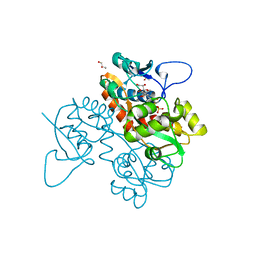 | | Structure of GluK1 ligand-binding domain in complex with N-(7-fluoro-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl)-2-hydroxybenzamide at 1.85 A resolution | | 分子名称: | CHLORIDE ION, GLYCEROL, Glutamate receptor ionotropic, ... | | 著者 | Kastrup, J.S, Frydenvang, K, Mollerud, S. | | 登録日 | 2018-03-14 | | 公開日 | 2019-01-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | N1-Substituted Quinoxaline-2,3-diones as Kainate Receptor Antagonists: X-ray Crystallography, Structure-Affinity Relationships, and in Vitro Pharmacology.
Acs Chem Neurosci, 10, 2019
|
|
5VZR
 
 | |
6CNW
 
 | |
6PXG
 
 | |
3K82
 
 | | Crystal Structure of the third PDZ domain of PSD-95 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Disks large homolog 4, GLYCEROL, ... | | 著者 | Camara-Artigas, A, Gavira, J.A. | | 登録日 | 2009-10-13 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Novel conformational aspects of the third PDZ domain of the neuronal post-synaptic density-95 protein revealed from two 1.4A X-ray structures
J.Struct.Biol., 170, 2010
|
|
5DW0
 
 | |
8AH7
 
 | |
5DW3
 
 | |
5DVZ
 
 | | Holo TrpB from Pyrococcus furiosus | | 分子名称: | PHOSPHATE ION, SODIUM ION, Tryptophan synthase beta chain 1 | | 著者 | Buller, A.R, Arnold, F.H. | | 登録日 | 2015-09-21 | | 公開日 | 2016-02-03 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Directed evolution of the tryptophan synthase beta-subunit for stand-alone function recapitulates allosteric activation.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8AH5
 
 | |
8AH6
 
 | |
8AH8
 
 | |
