7F8V
 
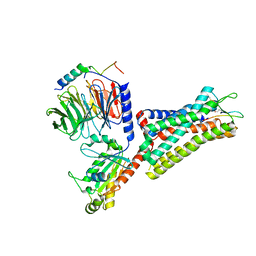 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | 分子名称: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-13 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | 分子名称: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8W
 
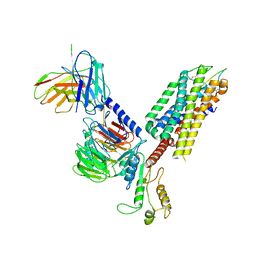 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | 分子名称: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-13 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8X
 
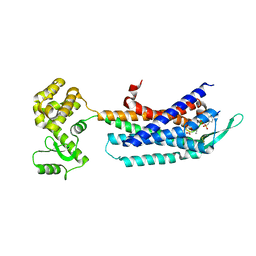 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with NN9056 | | 分子名称: | ASP-SMF-NLE-GLY-TRP-NLE-OEM-MEA-NH2 (NN9056), Cholecystokinin receptor type A,Endolysin | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | 分子名称: | Programmed cell death protein 1 | | 著者 | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-01-20 | | 公開日 | 2004-03-23 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
2B5Y
 
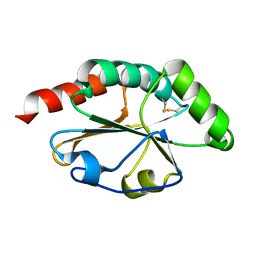 | |
2B5X
 
 | |
7EVN
 
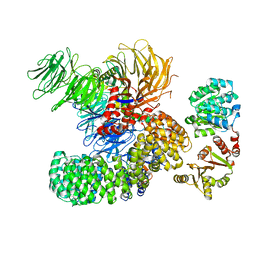 | | The cryo-EM structure of the DDX42-SF3b complex | | 分子名称: | ATP-dependent RNA helicase DDX42, PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, ... | | 著者 | Zhang, X, Zhan, X, Shi, Y. | | 登録日 | 2021-05-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural insights into branch site proofreading by human spliceosome
Nat.Struct.Mol.Biol., 2024
|
|
7EVO
 
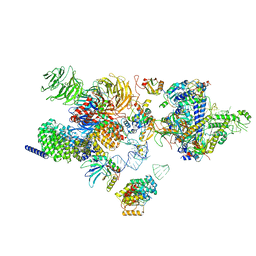 | | The cryo-EM structure of the human 17S U2 snRNP | | 分子名称: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, RNA helicase, ... | | 著者 | Zhang, X, Zhan, X, Shi, Y. | | 登録日 | 2021-05-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
5Z1V
 
 | | Crystal structure of AvrPib | | 分子名称: | AvrPib protein | | 著者 | Zhang, X, He, D, Zhao, Y.X, Taylor, I.A, Peng, Y.L, Yang, J, Liu, J.F. | | 登録日 | 2017-12-28 | | 公開日 | 2018-09-05 | | 最終更新日 | 2018-10-03 | | 実験手法 | X-RAY DIFFRACTION (1.661 Å) | | 主引用文献 | A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus.
Plant J., 96, 2018
|
|
5TEC
 
 | | Crystal structure of the TIR domain from the Arabidopsis thaliana NLR protein SNC1 | | 分子名称: | Protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 | | 著者 | Zhang, X, Bentham, A, Ve, T, Williams, S.J, Kobe, B. | | 登録日 | 2016-09-20 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Multiple functional self-association interfaces in plant TIR domains.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | 登録日 | 2010-04-14 | | 公開日 | 2011-04-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
6AHF
 
 | | CryoEM Reconstruction of Hsp104 N728A Hexamer | | 分子名称: | Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Zhang, X, Zhang, L, Zhang, S. | | 登録日 | 2018-08-17 | | 公開日 | 2019-02-13 | | 最終更新日 | 2019-04-10 | | 実験手法 | ELECTRON MICROSCOPY (6.78 Å) | | 主引用文献 | Heat shock protein 104 (HSP104) chaperones soluble Tau via a mechanism distinct from its disaggregase activity.
J. Biol. Chem., 294, 2019
|
|
2CSE
 
 | | Features of Reovirus Outer-Capsid Protein mu1 Revealed by Electron and Image Reconstruction of the virion at 7.0-A Resolution | | 分子名称: | Minor core protein lambda 3, Sigma 2 protein, guanylyltransferase, ... | | 著者 | Zhang, X, Ji, Y, Zhang, L, Harrison, S.C, Marinescu, D.C, Nibert, M.L, Baker, T.S. | | 登録日 | 2005-05-21 | | 公開日 | 2005-10-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution.
Structure, 13, 2005
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | 登録日 | 2017-12-14 | | 公開日 | 2018-08-08 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
5Z56
 
 | | cryo-EM structure of a human activated spliceosome (mature Bact) at 5.1 angstrom. | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z58
 
 | | Cryo-EM structure of a human activated spliceosome (early Bact) at 4.9 angstrom. | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z57
 
 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
6AH0
 
 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | 登録日 | 2018-08-15 | | 公開日 | 2018-11-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6AHD
 
 | | The Cryo-EM Structure of Human Pre-catalytic Spliceosome (B complex) at 3.8 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, Brr2, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | 登録日 | 2018-08-17 | | 公開日 | 2018-11-14 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
1F3L
 
 | |
8T3V
 
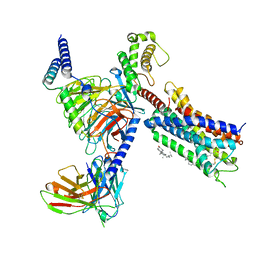 | | Cryo-EM structure of the DHA bound FFA1-Gq complex | | 分子名称: | CHOLESTEROL, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8SG1
 
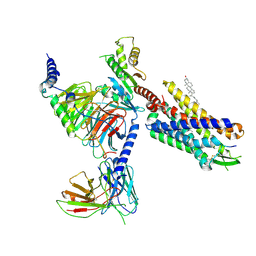 | | Cryo-EM structure of CMKLR1 signaling complex | | 分子名称: | CHOLESTEROL, Chemerin 9, Chemerin-like receptor 1, ... | | 著者 | Zhang, X, Zhang, C. | | 登録日 | 2023-04-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structural basis of G protein-Coupled receptor CMKLR1 activation and signaling induced by a chemerin-derived agonist.
Plos Biol., 21, 2023
|
|
8T3Q
 
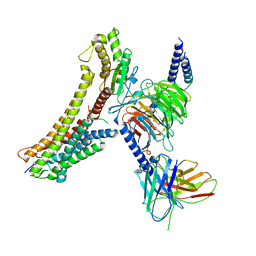 | | Cryo-EM structure of the DHA bound FFA4-Gq complex | | 分子名称: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3O
 
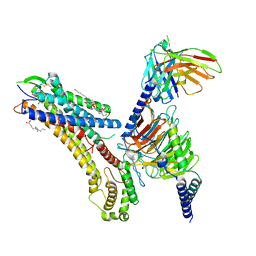 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | 分子名称: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | 著者 | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | 登録日 | 2023-06-07 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
