4QLI
 
 | |
4N6C
 
 | | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae. Northeast Structural Genomics Consortium (NESG) Target SpR36. | | 分子名称: | BROMIDE ION, uncharacterized protein | | 著者 | Vorobiev, S, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-10-11 | | 公開日 | 2013-10-30 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.548 Å) | | 主引用文献 | Crystal Structure of the B1RZQ2 protein from Streptococcus pneumoniae.
To be Published
|
|
4B50
 
 | |
4QI8
 
 | | Lytic polysaccharide monooxygenase 9F from Neurospora crassa, NcLPMO9F | | 分子名称: | COPPER (II) ION, Lytic polysaccharide monooxygenase, NITRATE ION | | 著者 | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | 登録日 | 2014-05-30 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
6RHV
 
 | | Crystal structure of mouse CD11b I-domain (CD11b-I) in complex with Staphylococcus aureus octameric bi-component leukocidin LukGH (LukH K319A mutant) | | 分子名称: | Beta-channel forming cytolysin, DIMETHYL SULFOXIDE, Integrin alpha-M, ... | | 著者 | Trstenjak, N, Milic, D, Djinovic-Carugo, K, Badarau, A. | | 登録日 | 2019-04-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Molecular mechanism of leukocidin GH-integrin CD11b/CD18 recognition and species specificity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8C5M
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with MTA | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Kremling, V, Sprenger, J, Oberthuer, D, Falke, S. | | 登録日 | 2023-01-09 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
4QI3
 
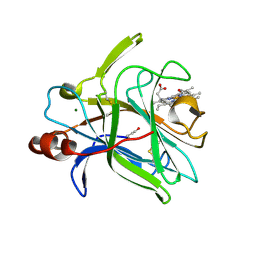 | | Cytochrome domain of Myriococcum thermophilum cellobiose dehydrogenase, MtCYT | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, MAGNESIUM ION, ... | | 著者 | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | 登録日 | 2014-05-30 | | 公開日 | 2015-07-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
3HVK
 
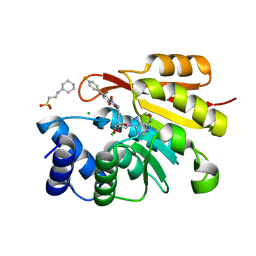 | | Rat catechol O-methyltransferase in complex with a catechol-type, purine-containing bisubstrate inhibitor - humanized form | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | 著者 | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | 登録日 | 2009-06-16 | | 公開日 | 2009-10-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
9MF5
 
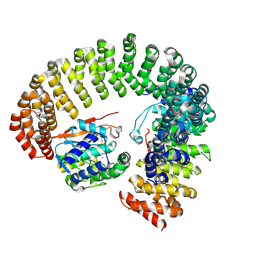 | |
8Q0S
 
 | | X-ray structure of the single chain monellin derivative MNEI | | 分子名称: | ACETATE ION, GLYCEROL, Monellin chain B,Monellin chain A, ... | | 著者 | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | 登録日 | 2023-07-29 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
8Q0R
 
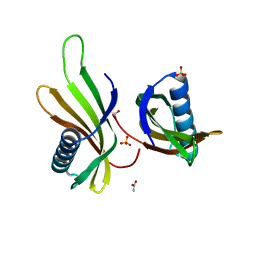 | | X-ray structure of MNEI mutant Mut9 (E23A, C41A, Y65R, S76Y) | | 分子名称: | ACETATE ION, Monellin chain B,Monellin chain A, SULFATE ION | | 著者 | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | 登録日 | 2023-07-29 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
9MIP
 
 | |
8QCI
 
 | | FCGBP D10 Assembly Segment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | 著者 | Yeshaya, N, Fass, D. | | 登録日 | 2023-08-27 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | VWD domain stabilization by autocatalytic Asp-Pro cleavage.
Protein Sci., 33, 2024
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | 分子名称: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | 著者 | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | 登録日 | 2014-06-27 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
8C7G
 
 | | Drosophila melanogaster Rab7 GEF complex Mon1-Ccz1-Bulli | | 分子名称: | Caffeine, calcium, zinc sensitivity 1, ... | | 著者 | Schaefer, J, Herrmann, E, Kuemmel, D, Moeller, A. | | 登録日 | 2023-01-15 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the metazoan Rab7 GEF complex Mon1-Ccz1-Bulli.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5EPI
 
 | |
6BNA
 
 | | BINDING OF AN ANTITUMOR DRUG TO DNA. NETROPSIN AND C-G-C-G-A-A-T-T-BRC-G-C-G | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(CBR)P*GP*CP*G)-3'), NETROPSIN | | 著者 | Kopka, M.L, Yoon, C, Goodsell, D, Pjura, P, Dickerson, R.E. | | 登録日 | 1984-08-30 | | 公開日 | 1984-10-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G.
J.Mol.Biol., 183, 1985
|
|
4PGJ
 
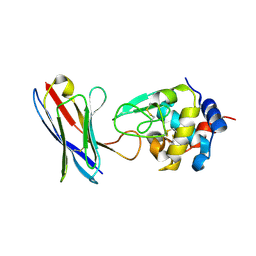 | |
6RUT
 
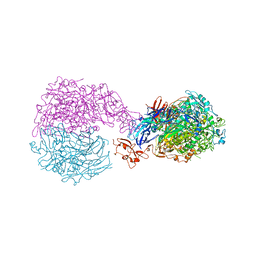 | |
8PVR
 
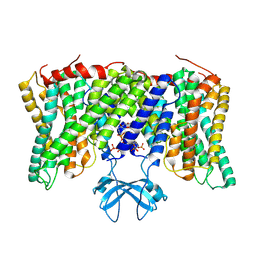 | | Cryo-EM structure of horse Nhe9 bound to PI(3,5)P2 | | 分子名称: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Sodium/hydrogen exchanger 9 | | 著者 | Kokane, S, Meier, P, Gulati, A, Delemotte, L, Drew, D. | | 登録日 | 2023-07-18 | | 公開日 | 2024-07-24 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | PIP 2 -mediated oligomerization of the endosomal sodium/proton exchanger NHE9.
Nat Commun, 16, 2025
|
|
6C2V
 
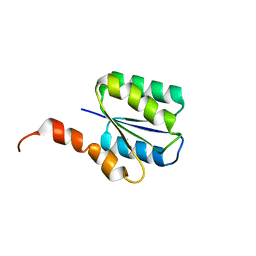 | | Solution structure of a phosphate-loop protein | | 分子名称: | phosphate-loop protein | | 著者 | Yang, F, Yang, W, Lin, Y.R, Romero Romero, M.L, Tawfik, D, Baker, D, Varani, G. | | 登録日 | 2018-01-09 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Simple yet functional phosphate-loop proteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8BXT
 
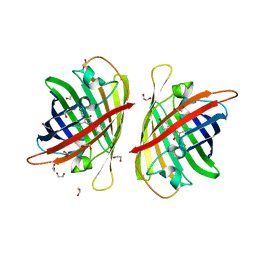 | | Structure of StayGold | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, StayGold | | 著者 | Ivorra-Molla, E, Akhuli, D, Crow, A. | | 登録日 | 2022-12-09 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A monomeric StayGold fluorescent protein.
Nat.Biotechnol., 42, 2024
|
|
7ZEG
 
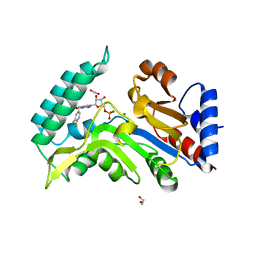 | | Human cytosolic 5' nucleotidase IIIB in complex with 3,4-diF-Bn7GMP | | 分子名称: | 1,2-ETHANEDIOL, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | 著者 | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | 登録日 | 2022-03-31 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
7ZEE
 
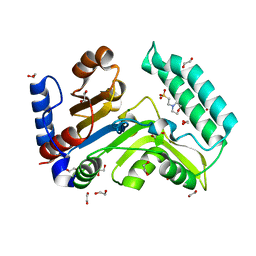 | | Human cytosolic 5' nucleotidase IIIB | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methylguanosine phosphate-specific 5'-nucleotidase, ... | | 著者 | Kubacka, D, Kozarski, M, Baranowski, M.R, Wojcik, R, Jemielity, J, Kowalska, J, Basquin, J. | | 登録日 | 2022-03-31 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Substrate-Based Design of Cytosolic Nucleotidase IIIB Inhibitors and Structural Insights into Inhibition Mechanism.
Pharmaceuticals, 15, 2022
|
|
8PXB
 
 | |
