4UXZ
 
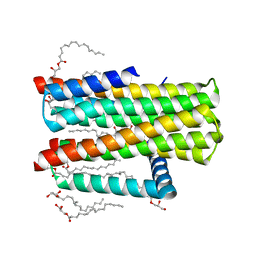 | | Structure of delta7-DgkA-syn in 7.9 MAG to 2.18 angstrom resolution | | 分子名称: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, ACETATE ION, ... | | 著者 | Li, D, Howe, N, Caffrey, M. | | 登録日 | 2014-08-27 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase.
Nat.Commun., 6, 2015
|
|
5KU3
 
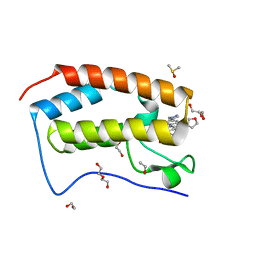 | | BRD4 bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | 分子名称: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, Bromodomain-containing protein 4, ... | | 著者 | Murray, J.M, Huang, W. | | 登録日 | 2016-07-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
8SZA
 
 | |
8SZB
 
 | |
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | 分子名称: | Endolysin,Rhodopsin,S-arrestin | | 著者 | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | 登録日 | 2015-08-28 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (7.7 Å) | | 主引用文献 | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
2XP2
 
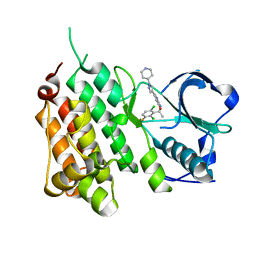 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with Crizotinib (PF-02341066) | | 分子名称: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, TYROSINE-PROTEIN KINASE RECEPTOR | | 著者 | McTigue, M, Deng, Y, Liu, W, Brooun, A, Timofeevski, S, Marrone, T, Cui, J.J. | | 登録日 | 2010-08-24 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure Based Drug Design of Crizotinib (Pf-02341066), a Potent and Selective Dual Inhibitor of Mesenchymal-Epithelial Transition Factor (C-met) Kinase and Anaplastic Lymphoma Kinase (Alk).
J.Med.Chem, 54, 2011
|
|
4ONC
 
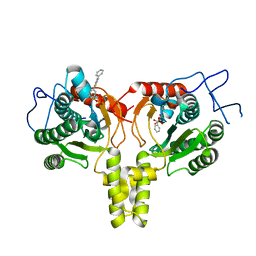 | | Crystal Structure of Mycobacterium Tuberculosis Decaprenyl Diphosphate Synthase in Complex with BPH-640 | | 分子名称: | Decaprenyl diphosphate synthase, [hydroxy(1,1':3',1''-terphenyl-3-yl)methanediyl]bis(phosphonic acid) | | 著者 | Feng, X, Chan, H.C, Ko, T.P. | | 登録日 | 2014-01-28 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structure and Inhibition of Tuberculosinol Synthase and Decaprenyl Diphosphate Synthase from Mycobacterium tuberculosis
J.Am.Chem.Soc., 136, 2014
|
|
8E80
 
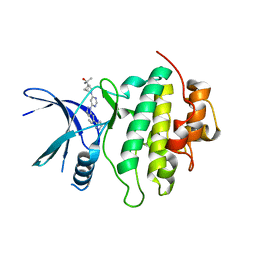 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 14 | | 分子名称: | 2-[(1r,4r)-2-{(6P)-6-[(6M)-6-(1H-pyrazol-5-yl)-1H-indazol-1-yl]pyrimidin-4-yl}-2-azabicyclo[2.1.1]hexan-4-yl]propan-2-ol, Serine/threonine-protein kinase Chk1 | | 著者 | Palte, R.L. | | 登録日 | 2022-08-25 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
8E81
 
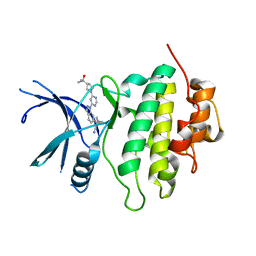 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 25 | | 分子名称: | (1S)-1-[(1P)-1-{6-[(3R)-3-(2-hydroxypropan-2-yl)pyrrolidin-1-yl]pyrimidin-4-yl}-1H-indazol-6-yl]spiro[2.2]pentane-1-carbonitrile, Serine/threonine-protein kinase Chk1 | | 著者 | Palte, R.L. | | 登録日 | 2022-08-25 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
6NHY
 
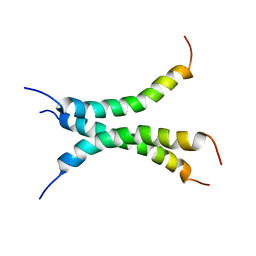 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | 分子名称: | Tumor necrosis factor receptor superfamily member 10B | | 著者 | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | 登録日 | 2018-12-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHW
 
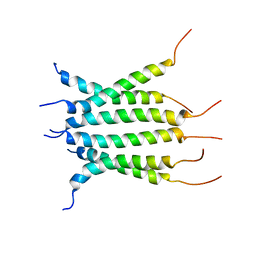 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | 分子名称: | Tumor necrosis factor receptor superfamily member 10B | | 著者 | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | 登録日 | 2018-12-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
7YJM
 
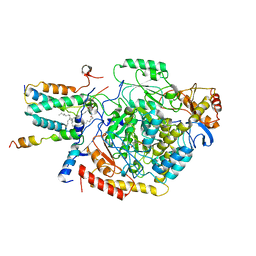 | | Cryo-EM structure of the monomeric atSPT-ORM1 complex | | 分子名称: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | 著者 | Xie, T, Liu, P, Gong, X. | | 登録日 | 2022-07-20 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJO
 
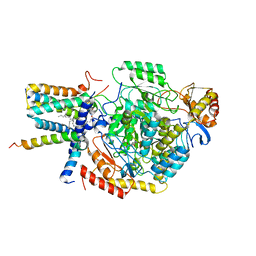 | | Cryo-EM structure of the monomeric atSPT-ORM1 (LCB2a-deltaN5) complex | | 分子名称: | Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORMDL family protein, ... | | 著者 | Xie, T, Liu, P, Gong, X. | | 登録日 | 2022-07-20 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJN
 
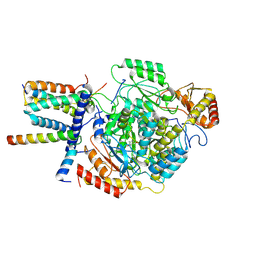 | | Cryo-EM structure of the monomeric atSPT-ORM1 (ORM1-N17A) complex | | 分子名称: | Long chain base biosynthesis protein 1, Long chain base biosynthesis protein 2a, ORMDL family protein, ... | | 著者 | Xie, T, Liu, P, Gong, X. | | 登録日 | 2022-07-20 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7YJK
 
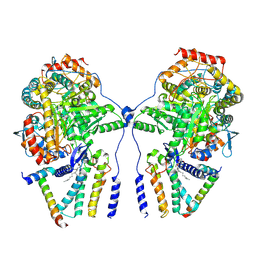 | | Cryo-EM structure of the dimeric atSPT-ORM1 complex | | 分子名称: | Long chain base biosynthesis protein 1, Long chain base biosynthesis protein 2a, N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ... | | 著者 | Xie, T, Liu, P, Gong, X. | | 登録日 | 2022-07-20 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanism of sphingolipid homeostasis revealed by structural analysis of Arabidopsis SPT-ORM1 complex.
Sci Adv, 9, 2023
|
|
7XY7
 
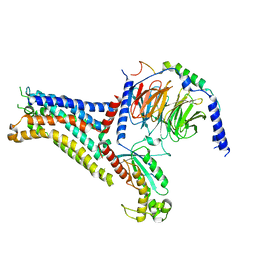 | | Adenosine receptor bound to a non-selective agonist in complex with a G protein obtained by cryo-EM | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | 登録日 | 2022-05-31 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
7XY6
 
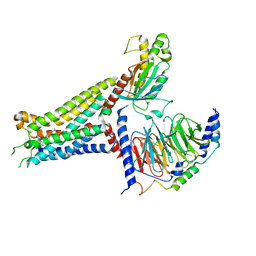 | | Adenosine receptor bound to an agonist in complex with G protein obtained by cryo-EM | | 分子名称: | 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | 登録日 | 2022-05-31 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
5O2F
 
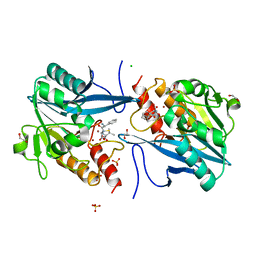 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | 登録日 | 2017-05-20 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5O2E
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed cefuroxime - new refinement | | 分子名称: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | 著者 | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | 登録日 | 2017-05-20 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5IE6
 
 | | Crystal structure of a lactonase mutant in complex with substrate b | | 分子名称: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | 著者 | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2016-02-25 | | 公開日 | 2017-01-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE5
 
 | | Crystal structure of a lactonase double mutant in complex with substrate a | | 分子名称: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | 著者 | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2016-02-25 | | 公開日 | 2017-01-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE4
 
 | | Crystal structure of a lactonase mutant in complex with substrate a | | 分子名称: | (3S,7R,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | 著者 | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2016-02-25 | | 公開日 | 2017-01-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
5IE7
 
 | | Crystal structure of a lactonase double mutant in complex with substrate b | | 分子名称: | (3S,7S,11E)-7,14,16-trihydroxy-3-methyl-3,4,5,6,7,8,9,10-octahydro-1H-2-benzoxacyclotetradecin-1-one, Zearalenone hydrolase | | 著者 | Zheng, Y.Y, Xu, Z.X, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2016-02-25 | | 公開日 | 2017-01-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Enhanced alph-Zearalenol Hydrolyzing Activity of a Mycoestrogen-Detoxifying Lactonase by Structure-Based Engineering
Acs Catalysis, 6, 2016
|
|
7Y8M
 
 | | Structure of ScIRED-R2-V3 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole | | 分子名称: | 2-[2,5-bis(fluoranyl)phenyl]pyrrolidine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, reductase | | 著者 | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | 登録日 | 2022-06-24 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
6JUJ
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | 分子名称: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | 登録日 | 2019-04-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.183 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
